Abstract
Advances in magnetic resonance imaging (MRI) and computational analysis technology have enabled comparisons among various primate brains in a three-dimensional electronic format. Results from comparative studies provide information about common features across primates and species-specific features of neuroanatomy. Investigation of various species of non-human primates is important for understanding such features, but the majority of comparative MRI studies have been based on experimental primates, such as common marmoset, macaques, and chimpanzee. A major obstacle has been the lack of a database that includes non-experimental primates’ brain MRIs. To facilitate scientific discoveries in the field of comparative neuroanatomy and brain evolution, we launched a collaborative project to develop an open-resource repository of non-human primate brain images obtained using ex vivo MRI. As an initial open resource, here we release a collection of structural MRI and diffusion tensor images obtained from 12 species: pygmy marmoset, owl monkey, white-fronted capuchin, crab-eating macaque, Japanese macaque, bonnet macaque, toque macaque, Sykes’ monkey, red-tailed monkey, Schmidt’s guenon, de Brazza’s guenon, and lar gibbon. Sixteen postmortem brain samples from the 12 species, stored in the Japan Monkey Centre (JMC), were scanned using a 9.4-T MRI scanner and made available through the JMC collaborative research program (http://www.j-monkey.jp/BIR/index_e.html). The expected significant contributions of the JMC Primates Brain Imaging Repository include (1) resources for comparative neuroscience research, (2) preservation of various primate brains, including those of endangered species, in a permanent digital form, (3) resources with higher resolution for identifying neuroanatomical features, compared to previous MRI atlases, (4) resources for optimizing methods of scanning large fixed brains, and (5) references for veterinary neuroradiology. User-initiated research projects beyond these contributions are also anticipated.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the quest for a scientific understanding of the nature of humans, it is crucial to elucidate the evolutionary process of the special features of the central nervous system in modern humans (Homo sapiens) (Preuss 2011; Rilling 2014). The changes in the brain during the evolution from early primates to modern humans involved a marked increase of the overall brain size, and especially of the size of the neocortex (Roth and Dicke 2005; Dunbar and Shultz 2017; Kaas 2017).
One limitation of fossil-based studies is their lack of information about brain structures (Holloway et al. 2004; Hofman and Falk 2012). To further investigate changes of various component brain structures that occurred during human evolution, comparative neuroanatomy, in which similarities and differences among human and non-human primate brains are identified, is necessary. This approach provides information about the common features among primates, the specific features of each primate, and the human-specific features, of the brain, which are all necessary to understand the evolutionary changes of the brain structures and functions, including those specifically seen in humans, such as language function and mental disorders.
Traditional approaches and their limitations for primate comparative neuroanatomy
Among various measures of comparative neuroanatomy, volume, shape, gyrification, cytoarchitecture, and the connectivity among cortical areas are often used as the targets of research (Krubitzer and Kaas 2005). There are macroscopic and microscopic approaches for making the respective measurements.
Macroscopic approaches have been used to investigate the gross anatomical features such as the size of the brain areas and sulcal anatomy. This approach has proven to be successful for examining the relationship between gray and white matter of the cerebral cortex (Zhang and Sejnowski 2000) and for investigating the subcortical area (Barton and Harvey 2000; Barton 2006).
Microscopic approaches allow us to study informative details of the features of brain anatomy such as cytoarchitecture, myelination, and connectivity of the brain regions (Morecraft et al. 2009). These methods represent the mainstream in comparing brain structure among species, and they are continuously being refined (Markov et al. 2012; Mars et al. 2014).
However, it is very laborious and expensive to use the comparative neuroanatomy approach. Although recent studies have continued to use such primate comparative neuroanatomy employing various tracing methods, the cost and time investment required mean that this approach is not often applied to more than four or five species in a study (Mars et al. 2014). Recently, a database of the axonal projections and the connectivity of the whole brain in macaque monkeys was released for the public as an online resource (CoCoMac, http://cocomac.g-node.org/main/index.php), but the microscopic features of the brain have not been fully explored in other non-human primates.
Major limitations of histological studies include the difficulty of re-analysis and of preservation without degradation of specimens. Once a brain has been cut and stained, it cannot be cut and stained again in other ways. Brain samples that are immersed in formalin for a long time suffer from acidity, dehydration (Shepherd et al. 2009; Shatil et al. 2016), and degradation of the proteins (D’Arceuil and de Crespigny 2007). Therefore, formalin fixation of specimens is not an ideal way to store specimens permanently for use in obtaining neuroanatomical information about the primate brain.
Advances in ex vivo MRI and their limitations for primate comparative neuroanatomy
During the past two decades, advances in MRI technology and computational imaging analysis technology have made it possible to conduct direct comparisons of the brain organization of the human with that of non-human primate model animals, for example, common marmosets (Callithrix jacchus), rhesus macaques (Macaca mulatta), and chimpanzees (Pan troglodytes).
The major advantages of MRI for comparative neuroanatomy are as follows. First, MRI can be used to quantify various macroscopic and microscopic characteristics of the brain organization. Second, MRI allows us to collect quantitative whole-brain images within the relatively short time required for image acquisition compared to histological examination (which requires, for example, paraffin embedding, slicing, sectioning, mounting, deparaffinization, antigen activation, staining, and visualization using a microscope). Finally, as their greatest advantage, MRI techniques are non-destructive. Further details of primate comparative neuroimaging using MRI were reviewed by Rilling (2014) and Mars et al. (2014).
In traditional neuroanatomical approaches, in order to investigate some particular area of the brain using stains or tracers or viruses, it is necessary to cut the brain into slices or to block some regions irreversibly. In MRI approaches, this problem can be addressed simply by scanning the same whole brain repeatedly, which allows us to acquire information about the various brain tissues and components by using different scanning protocols.
Its non-destructive properties make MRI an ideal method for studying postmortem (ex vivo) primate brains. Compared to in vivo scanning, ex vivo imaging allows for extremely long MRI experiments that are free of subject motion and other sources of physiological noise (Shatil et al. 2016). Therefore, ex vivo MRI produces higher spatial resolution and signal-to-noise ratios than are achievable by in vivo MRI.
In comparative neuroanatomical imaging, structural MRI has been used to compare the brain’s macroscopic anatomical proportions (e.g., tissue volume, sulci, and gyri). Diffusion MRI has been used to compare microstructures (e.g., expansion of the isocortex) and white matter connectivity. In the next paragraph, we introduce some representative findings obtained previously by comparative neuroanatomical imaging using structural and diffusion MRI approaches with primate postmortem brain samples, and explain the advantages and limitations of these imaging approaches.
The structural ex vivo MRI approach is useful for investigating fetal brain development in not only humans but also non-human primates. Some non-human primate MRI research groups defined the developmental patterns of the cerebral sulci and gyri during fetal stages in crab-eating macaque (Macaca fascicularis) and common marmoset (Sawada et al. 2009, 2012; Hikishima et al. 2013; Sawada et al. 2014). Furthermore, they clarified the similarities and differences of gyrification patterns between humans and non-human primates by comparing the patterns of the non-human primates with those of the human fetus (Sawada et al. 2009, 2012; Hikishima et al. 2013; Sawada et al. 2014).
Diffusion MRI measures water diffusion as a probe to examine the microscopic organization of anatomical structures within the anatomical region represented by each pixel (Basser et al. 1994a, b). One of the major strengths of diffusion MRI is its capability of delineating well-aligned structures, such as the white matter bundles or columnar organization of the cortex. It can also delineate trajectories of major white matter bundles three-dimensionally (Mori and van Zijl 2002). It has been demonstrated that white matter bundles delineated by diffusion MRI are comparable to those delineated by histology in the rhesus macaques (Schmahmann et al. 2007).
The diffusion ex vivo MRI approach has been used to investigate similarities and differences between brains of humans and non-human primates. Comparative diffusion MRI studies showed that the trajectories of the major U-fibers and the extreme capsule are similar between humans and macaques (Oishi et al. 2011), whereas the trajectory of the left arcuate fasciculus, which is related to language, is different in humans compared to that of chimpanzees or macaques (Rilling et al. 2008). A recent comparative diffusion MRI study revealed that an increase of cross-cortical connections emerged in the primate lineage after the split between primates and other mammals (Charvet et al. 2017). Diffusion MRI is also useful for quantification of developmental changes in cytoarchitecture. For example, cortical layer-specific developmental changes were delineated by diffusion anisotropy in baboon brains, the pattern of which was comparable to that in rats and humans (Kroenke et al. 2005).
Although ex vivo MRI approaches allow researchers to collect detailed information about brain anatomy, previous imaging studies focused only on experimental primate models. To better understand anatomical features and connectivity of the brain across the primate lineage, more species need to be studied. Involvement of researchers outside the primate research community is also important, as indicated by the fact that human brain MRI studies have been strongly driven by computer scientists, statisticians, and computational neuroanatomists. Notably, the major current barrier against comparative brain MRI studies in primates is the lack of an ex vivo non-human primate MRI database that includes a variety of non-human primate species and is freely accessible to researchers who lack MRI resources and animal facilities. The brain MRI databases currently available are a chimpanzee database [The National Chimpanzee Brain Resource (NCBR), http://www.chimpanzeebrain.org/] and a macaque monkey database (The PRIMate Data Exchange, http://fcon_1000.projects.nitrc.org/indi/indiPRIME.html). To date, there are no other MRI databases of other non-human primate species.
Aims of the Japan Monkey Centre Primates Brain Imaging Repository
To advance research in the field of comparative primate neuroanatomy, we launched a project to create a brain MRI repository that consists of MRI data of various non-human primate brains. In order to include non-experimental primates, we used formalin-fixed brains collected from primates that had lived in the Japan Monkey Centre (JMC), which houses the largest number of non-human primate species in one facility in the world. Since the foundation of the JMC in 1956, more than 2400 postmortem brain samples have been collected there from more than 100 non-human primate species, including endangered species such as gibbons (Hylobatidae), gorillas (Gorilla), chimpanzees, and vulnerable species such as owl monkeys (Aotus trivirgatus).
This primate brain sample collection is advantageous for our secondary goal of permanently archiving information about endangered species. It has been reported that about 60% of non-human primate species are now threatened with extinction. Not only members of the hominoid family, such as gibbons, orangutans (Pongo), gorillas, and chimpanzees, but also all 16 extant primate families are threatened with extinction (Estrada et al. 2017). Furthermore, the populations of 75% of primate species are decreasing globally (Estrada et al. 2017). Therefore, recording and keeping the information about these species permanently in electronic form is of great importance.
In this study, as a first step, we acquired structural MRI and DTI (diffusion tensor imaging) data from 16 primate brain samples (12 species) that are part of the JMC collection. Then we used these data to develop the JMC Primates Brain Imaging Repository (http://www.j-monkey.jp/BIR/index_e.html) in order to make the electronic data available to the research community.
Methods
The Japan Monkey Centre primate fixed brain collection
We used 16 postmortem samples of non-human primate brains (12 species), under protocols approved by the JMC’s review board for primate research (permits #2014013, #2015019, and #2016017). Details about postmortem brain specimens are given in Table 1. The brains were immersed in 10% neutral buffered formalin, and fixed for more than 1 month. After the fixation, some of the brain samples were immersed in phosphate-buffered saline with 1 mM gadopentetate dimeglumine (Magnevist, Berlex Imaging, Wayne, NJ, USA) (GD-PBS) for at least 2 weeks before the scan to enhance image contrast. The rest of the scans were performed without contrast enhancement, as detailed in the Discussion section (Tables 2, 3). The samples were placed in a customized polyethylene container that was filled with Fluorinert (Fluorinert™ FC-43, 3 M Japan Limited, Tokyo, Japan) to prevent dehydration and reduce susceptibility to artifacts at tissue margins. The temperature of the specimens was maintained at 24 °C during the scans in accordance with the International Industrial Standard (IEC 60601-2-33) (Bottomley et al. 1984; Fix et al. 2000).
Image acquisition
The whole-brain scans were performed using a 9.4-T Biospec 90/20 MRI scanner (Bruker Biospin GmbH; Ettlingen, Germany) with a triple-axis gradient system (maximum gradient strength of 300 mT/m), using a transmitting and receiving two-channel quadrature coil with an 86-mm inner diameter at Jikei University. T2-weighted images were acquired using a three-dimensional rapid acquisition with refocused echoes (RARE) sequence. DTIs were acquired using a conventional pulsed gradient spin echo (PGSE) sequence. The acquisition parameters and the lengths of the scan time for T2-weighted images and DTIs are shown in Tables 2 and 3, respectively.
Image processing
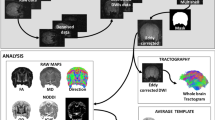
Figure 1 shows a schematic of the image processing. T2-weighted images were processed using the following series of procedures using ANALYZE v. 9.0 software (Mayo Clinic, Mayo Foundation, Rochester, MN, USA) and DtiStudio (https://www.mristudio.org) (Jiang et al. 2006), and DiffeoMap (https://www.mristudio.org) (Fig. 1). (1) T2-weighted images were resampled to isotopic voxels. Image resolution varied depending on the species, ranging from 0.15 to 0.25 mm. (2) Resampled T2-weighted images were aligned to a standard anatomical orientation with the transaxial plane parallel to the anterior commissure-posterior commissure (AC-PC) line and perpendicular to the interhemispheric fissure. (3) A radio-frequency bias field correction was applied using the Bias Correction function implemented in DiffeoMap.
Schematic of image processing in this database. T2-weighted images were processed firstly by performing resampling to isotopic voxel size, followed by bias field correction and rigid alignment to AC-PC line. The DTIs were processed firstly by performing tensor calculation, followed by creation of DTI maps. Finally, the DTIs and DTI maps were coregistered to the AC-PC aligned T2-weighted image
DTIs of each brain were analyzed using the following series of procedures using DtiStudio (Pierpaoli et al. 2001; Jiang et al. 2006; Mori and Zhang 2006) and DiffeoMap (Fig. 1). (1) The eigenvalues and associated eigenvectors (Basser et al. 1994b) of the voxel-wise diffusion tensors were computed at each pixel along with the apparent diffusion coefficient (ADC), fractional anisotropy (FA), primary eigenvector, axial diffusivity (λ|, the primary eigenvalue), and radial diffusivity (λ⊥, the average of the secondary and tertiary eigenvalues). (2) The mean diffusion-weighted image (DWI) was generated by averaging all diffusion-weighted images. (3) The color-coded orientation map (color map) was created by assigning red, green, and blue components equal to the ratio of the absolute values of x (medial–lateral), y (anterior–posterior) and z (superior–inferior) components of the primary eigenvector, where intensity was proportional to FA (Makris et al. 1997; Pajevic and Pierpaoli 1999). (4) DWI, FA, and color maps for each sample were then resliced to the isotopic voxel identical to that of the corresponding T2-weighted image. (5) The least diffusion-weighted image (b0) of the DTI was transformed to the T2-weighted image using six-parameter rigid transformation of the Automatized Image Registration (AIR) (Woods et al. 1998) implemented in DiffeoMap, and the transformation was applied to the corresponding DWI and tensor field, from which FA and color maps were recalculated. (6) Finally, all images were saved in the ANALYZE format.
The T2-weighted images and DTIs were quality controlled by the authors, and those with poor quality were re-scanned or excluded from the repository.
Results
We succeeded in acquiring high-spatial-resolution T2-weighted images and DTIs (DWIs, b0, FA map, and color map) from various non-human primate brain samples (Figs. 2, 3, 4, 5, 6, 7, 8). A histological atlas of the rhesus macaque brain (Paxinos et al. 2000) was used to identify the anatomical features. A DTI atlas of the human brain (Mori et al. 2005) was used to identify white matter fibers. The details of the results were as follows.
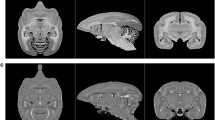
Representative samples of brain surfaces of 12 non-human primate brain samples in the database. The images are surface reconstructions of the brain for each brain sample in the following 12 non-human primate species: a pygmy marmoset (Pr6324), b owl monkey (Pr6396), c white-fronted capuchin (Pr6365), d crab-eating macaque (Pr6413), e Japanese macaque (Pr6378), f bonnet macaque (Pr6375), g toque macaque (Pr6182), h Sykes’ monkey (Pr6443), i red-tailed monkey (Pr6379), j Schmidt’s guenon (Pr6002), k de Brazza’s guenon (Pr6230), and l lar gibbon (Pr5972). The left image shows a photograph of the brain samples and the right image shows a surface reconstruction of the T2-weighted image for each brain sample
Representative samples of T2-weighted images of 12 non-human primate brain samples in the database. The images are T2-weighted images from the mid-axial, mid-coronal, and almost mid-sagittal planes, for each brain sample in the following 12 non-human primate species: a pygmy marmoset (Pr6324), b owl monkey (Pr6396), c white-fronted capuchin (Pr6365), d crab-eating macaque (Pr6413), e Japanese macaque (Pr6378), f bonnet macaque (Pr6375), g toque macaque (Pr6182), h Sykes’ monkey (Pr6443), i red-tailed monkey (Pr6379), j Schmidt’s guenon (Pr6002), k de Brazza’s guenon (Pr6230), and l lar gibbon (Pr5972)
Representative samples of the DTI images of 11 non-human primate brain samples in the database. The images are DWIs, b0, FA and color maps from the mid-axial plane for each brain sample in the following 11 non-human primate species: a pygmy marmoset (Pr6324), b owl monkey (Pr6396), c white-fronted capuchin (Pr6365), d crab-eating macaque (Pr6413), e Japanese macaque (Pr6378), f bonnet macaque (Pr6375), g toque macaque (Pr6182), h Sykes’ monkey (Pr6443), i red-tailed monkey (Pr6379), j Schmidt’s guenon (Pr6002), and k de Brazza’s guenon (Pr6230). Red, green, and blue indicate fiber orientations along the medial–lateral, anterior–posterior, and superior–inferior axes, respectively
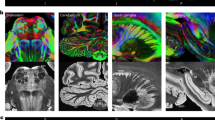
Demonstration of the neuroanatomical details resolved in the brain surface. The image shows lateral (top) and dorsal (bottom) views of the brain surface from T2-weighted images in a pygmy marmoset (Pr6324), b Sykes’ monkey (Pr6443), and c Schmidt’s guenon (Pr6002). Neuroanatomical abbreviations in this MRI database are represented in accordance with the histological atlas of the rhesus macaque brain (Paxinos et al. 2000). The abbreviations for features in the brain surface are: AG angular gyrus, ASPD anterior supraprincipal dimple, ASPG anterior superior parietal gyrus, CG cingulate gyrus, CS central sulcus, ECAL external calcarine sulcus, IAR inferior arcuate sulcus, IFG inferior frontal gyrus, IOS inferior occipital sulcus, IPD infraprincipal dimple, IPS intraparietal sulcus, LF lateral fissure, LU lunate sulcus, MFG middle frontal gyrus, MG marginal gyrus, MTG middle temporal gyrus, OG occipital gyrus, OTG occipito-temporal gyrus, POS parietooccipital sulcus, PRCG precentral gyrus, PSPG posterior superior parietal gyrus, SAR superior arcuate sulcus, SFG superior frontal gyrus, SPCD superior precentral dimple, STG superior temporal gyrus, STS superior temporal sulcus, SU superior postcentral dimple, TPG temporal polar gyrus
Demonstration of the neuroanatomical details resolved with the high-spatial-resolution T2-weighted image and DTI color map from the mid-axial plane. The image shows T2-weighted images (middle column) and color maps (right column) from the mid-axial plane in a pygmy marmoset (Pr6324), b Sykes’ monkey (Pr6443), and c Schmidt’s guenon (Pr6002). Red, green, and blue indicate fiber orientations along the medial–lateral, anterior–posterior, and superior–inferior axes, respectively. Neuroanatomical abbreviations in this MRI database are represented in accordance with the histological atlas of the rhesus macaque brain (Paxinos et al. 2000). The abbreviations for features in T2-weighted images are: ACB accumbens, CD caudate nucleus, EGP external globus pallidus, HIP hippocampus, IGP internal globus pallidus, GN geniculate nucleus, IPUL inferior pulvinar, MT mammillothalamic tract, PAG periaqueductal gray, PU putamen, SC superior colliculus, THAL thalamus. The abbreviations for features in color maps are: AC anterior commissure, CG cingulum, EC external capsule, FX fornix, FX-C column of the fornix, FX-P precommissural part of the fornix, PLIC posterior limb of the internal capsule, SS sagittal stratum, ST stria terminalis
Demonstration of the neuroanatomical details resolved with the high-spatial-resolution T2-weighted image and DTI color map from mid-coronal plane. The image shows T2-weighted images (middle column) and color maps (right column) from the mid-coronal plane in a pygmy marmoset (Pr6324), b Sykes’ monkey (Pr6443), and c Schmidt’s guenon (Pr6002). Red, green, and blue indicate fiber orientations along the medial–lateral, anterior–posterior, and superior–inferior axes, respectively. Neuroanatomical abbreviations in this MRI database are represented in accordance with the histological atlas of the rhesus macaque brain (Paxinos et al. 2000). The abbreviations for features in T2-weighted images are: AM anteromedial thalamic nucleus, AV anteroventral thalamic nucleus, CD caudate nucleus, CL claustrum, EGP external globus pallidus, GP globus pallidus, HIP hippocampus, IGP internal globus pallidus, PC paracentralis thalamic nucleus, PU putamen, R reticularis thalamic nucleus, RE reuniens thalamic nucleus, SNR substantia nigra, STH subthalamic nucleus, THAL thalamus, VA ventralis thalamic anterior nucleus, VL ventralis thalamic lateralis nucleus. The abbreviations for features in color maps are: CC corpus callosum, CG cingulate, CP cerebral peduncle, EC external capsule, FX fornix, OPT optic tract, PLIC posterior limb of the internal capsule, SFO superior fronto-occipital fasciculus, ST stria terminalis
Demonstration of the neuroanatomical details resolved with the high-spatial-resolution T2-weighted image and DTI color map from almost mid-sagittal plane. The image shows T2-weighted images (middle column) and color maps (right column) from almost mid-sagittal plane in a pygmy marmoset (Pr6324), b Sykes’ monkey (Pr6443), and c Schmidt’s guenon (Pr6002). Red, green, and blue indicate fiber orientations along the medial–lateral, anterior–posterior, and superior–inferior axes, respectively. Neuroanatomical abbreviations in this MRI database are represented in accordance with the histological atlas of the rhesus macaque brain (Paxinos et al. 2000). The abbreviations for features in T2-weighted images are: CB1 cerebellar lobule 1, CB2 cerebellar lobule 2, CB3 cerebellar lobule 3, CB4 cerebellar lobule 4, CB5 cerebellar lobule 5, CB6 cerebellar lobule 6, CB7 cerebellar lobule 7, CB8 cerebellar lobule 8, CB9 cerebellar lobule 9; CB10 cerebellar lobule 10. The abbreviations in color maps are: AC anterior commissure, CC corpus callosum, CG cingulum, DSCP decussation of the superior cerebellar peduncles, FX fornix, GCC genu of the corpus callosum, MCP middle cerebellar peduncle, OPT optical tract, PCT pontocerebellar tract, SCC splenium of the corpus callosum
T2-weighted images
Representative examples of the T2-weighted images are shown in Figs. 2, 3, 5, 6, 7, and 8. The sulci and gyri were identified in the brain surface of T2-weighted images. For example, in the lateral view of the Sykes’s monkey (Pr 6443) brain, the infraprincipal dimple, inferior arcuate sulcus, central sulcus, intraparietal sulcus, lateral fissure, superior temporal sulcus, lunate sulcus, inferior occipital sulcus, and external calcarine sulcus were identified. The middle temporal gyrus and temporal polar gyrus were also delineated. In the dorsal view of the brain, the anterior supraprincipal dimple, superior arcuate sulcus, superior precentral dimple, central sulcus, intraparietal sulcus, superior postcentral dimple, superior temporal sulcus, and parieto-occipital sulcus were identified. The superior frontal gyrus, middle frontal gyrus, inferior frontal gyrus, precentral gyrus, anterior superior parietal gyrus, cingulate gyrus, posterior superior parietal gyrus, angular gyrus, marginal gyrus, occipito-temporal gyrus, superior frontal gyrus, and occipital gyrus were also delineated (Fig. 5).
The T2-weighted contrast enables detailed anatomical delineation in the deep brain regions. For example, in Sykes’s monkey (Pr6443), the T2-weighted image provided striking contrasts that made it possible to delineate the accumbens, basal ganglia (putamen, external globus pallidus, and internal globus pallidus), thalamus, mammillothalamic tract, periaqueductal gray, pulvinar, caudate nucleus, hippocampus, and superior colliculus from the mid-axial plane (Fig. 6). At the mid-coronal plane, the caudate nucleus, thalamic nucleus, and basal ganglia were identified (Fig. 7). Nearly adjacent to the mid-sagittal plane, ten cerebellar lobules were distinctly delineated (Fig. 8).
Some images included image artifacts caused by small incisions on the brain surface (Fig. 9) or air bubbles (Fig. 10a). Moreover, concentric contrast inhomogeneities were observed in large brain samples such as those of Japanese macaque and lar gibbons (Fig. 11a). However, contrast inhomogeneity was not observed in the small brain samples. Since 2017, we have scanned brain samples without GD-PBS, and can acquire brain images with good contrast for large brain samples (Fig. 11b).
Representative sample of image artifacts caused by air bubbles and granular representation. The image shows image artifacts caused by air bubbles and granular representation in Japanese macaque (Pr6158). a T2-weighted images and b color maps. The axial, coronal, and sagittal planes are indicated. Red, green, and blue indicate fiber orientations along the medial–lateral, anterior–posterior, and superior–inferior axes, respectively. The red arrows indicate air bubbles. The yellow arrows indicate the granular representation, probably due to degradation of the brain tissue
Representative sample of an image artifact caused by concentric contrast inhomogeneities with GD-PBS. The image shows the concentric contrast inhomogeneities observed with GD-PBS. a T2-weighted images of a Japanese macaque brain sample (Pr6378) preconditioned by GD-PBS and b T2-weighted images of a Bonnet Macaque sample (Pr6375) without GD-PBS preconditioning. The axial, coronal, and sagittal planes are indicated. The concentric contrast inhomogeneities (red arrows) are seen in a but not in b
Diffusion tensor images
Representative scalar maps derived from DTIs are shown in Figs. 4, 6, 7, and 8. Structural orientation-based contrast in the color maps enabled us to delineate well-aligned structures, such as white matter bundles and columnar structures within the cortex. For example, in Sykes’s monkey (Pr6443), the color maps provided striking contrasts that made it possible to delineate the anterior commissure, external capsule, posterior limb of the internal capsule, sagittal stratum, column of the fornix, precommissural part of the fornix, fornix, stria terminalis, and cingulum at the mid-axial plane (Fig. 6). At the mid-coronal plane, the posterior limb of the internal capsule, external capsule, cerebral peduncle, optic tract, corpus callosum, cingulate, fornix, superior fronto-occipital fasciculus, and stria terminalis were identified (Fig. 7). Close to the mid-sagittal plane, the anterior commissure, optical tract, decussation of the superior cerebellar peduncles, pontocerebellar tract, corpus callosum, genu of the corpus callosum, splenium of the corpus callosum, cingulum, fornix, and middle cerebellar peduncle were distinctly delineated (Fig. 8).
In addition to the image artifacts caused by air bubbles, there were some images with a color map demonstrating granular representation, probably due to degradation of the brain tissue (Fig. 10b).
Incidental finding
The brain sample of the red-tailed monkey (Pr6379) included a space-occupying lesion that was observed to extend into the right frontal lobe, parietal lobe, temporal lobe, basal ganglia, internal capsule, and thalamus in the T2-weighted images (Fig. 12a). We also observed that a low signal in the same area in the DWIs and color maps (Fig. 12b, c). However, the boundary of the space-occupying lesion was unclear (Fig. 12a, b–c). This lesion was not visible from the brain surface, and therefore was not recognized until the brain MRI scan was performed. Of note, the brain surface was clear and looked normal (Fig. 2i). The monkey was thought to have died of old age since a gross anatomical observation could not identify a specific cause of death.
Images of a brain with a space-occupying lesion. The images show T2-weighted images, DWIs, and color maps in a red-tailed monkey (Pr6379). a T2-weighted images, b DWIs, and c color maps. The axial, coronal, and sagittal planes are indicated. Areas surrounded by red lines represent the space-occupying lesion. Red, green, and blue indicate fiber orientations along the medial–lateral, anterior–posterior, and superior–inferior axes, respectively. The upper rows show the original images and the lower rows show the images with the space-occupying lesion surrounded by a red line
Discussion
Significance of the Japan Monkey Centre Primates Brain Imaging Repository
Toward our long-term goal of establishing the JMC Primates Brain Imaging Repository, which will include images from more than 100 non-human primate species, MRIs from 12 species were released as the first issue. High-resolution T2-weighted images and DTIs were obtained by scanning with an ultra-high magnetic field scanner, and the images are now open to the research community through our website.
We expect fifth significant contributions of our repository. First, our imaging database provides a resource for comparative neuroscience to understand the brain morphology and neuronal connectivity of each species. Moreover, metadata such as body weight and brain weight are also provided. This information is needed to calculate the “encephalization quotient”, EQ = Ea/Ee, which indicates the extent to which the brain size of a given species, Ea, deviates from the expected brain size, Ee, based on a “standard” for mammals (Jerison 1973). The EQ ranking is considered to predict intelligence (Jerison 1973). Thus, users of this database can investigate the relationship between EQ and the absolute or relative size of the cerebral cortex to find better predictors for high-order cognitive abilities among primates.
We also anticipate that this imaging repository will facilitate discoveries in the fields of evolutionary and developmental neuroscience. We encourage its use by researchers that belong to research fields outside traditional primatology, such as computer science, mathematical modeling, and medicine.
Second, the JMC Primates Brain Imaging Repository will be a permanent repository of primate brains, since the images are stored as digital data. Unlike fixed brains, digital data are not subject to deterioration, and can be distributed worldwide. The images can be analyzed unlimitedly in various ways. This repository is especially important for preserving information about endangered species, since without this repository, this precious information would be lost. We will acquire additional whole-brain MRIs from endangered great apes, such as orangutans, gorillas, and chimpanzees, in the near future.
Third, our repository provides brain MRIs of non-human primates with higher resolution compared to the previous MRI atlases (Okano and Nomura 2008; Saleem and Logothetis 2012). These previous atlases showed the anatomical features on MRIs, combined with histological atlases in common marmosets and rhesus macaques based on Klüver–Barrera staining and immunohistochemical staining (Okano and Nomura 2008; Saleem and Logothetis 2012). Although we do not have the histological data corresponding to the MRI data of the non-human primate brain samples in this database, we found that our high-resolution MRIs enable us to delineate highly detailed anatomical features in non-human primates, especially white matter bundles. For example, we are able to identify nine white matter bundles (anterior commissure, cingulum, external capsule, fornix, column of the fornix, precommissural part of the fornix, posterior limb of the internal capsule, sagittal stratum, and stria terminalis) in the mid-axial plane on our non-human primate MRIs. This is an advance compared to the previous MRI atlas of the common marmoset, which defined only five white matter bundles (anterior commissure, posterior commissure, fornix, internal capsule, and sagittal stratum) in the mid-axial plane (Okano and Nomura 2008). The previous MRI atlas of the rhesus macaque did not show white matter bundles (Saleem and Logothetis 2012).
Fourth, the repository also provides opportunities to optimize protocols for postmortem brain scans. Although we tried as far as possible to apply state-of-the-art scanning protocols to acquire the ex vivo MRIs, there still remains room to further improve the image quality. The information about the scanners and the scan protocols are fully open. This provides opportunities for MR physicists to develop novel MRI sequences optimized for primate brain samples of various sizes, or for researchers who are interested in image post-processing to develop novel image analysis methods. Our database will serve as a resource for such new scan protocols or analysis methods.
Finally, the repository has the potential to serve as a disease reference. During the process of data accumulation in the future, we expect the inclusion of diseased brains. In the current data series, a space-occupying lesion was found incidentally in the right hemisphere of a red-tailed monkey. We plan to investigate its pathology in the future. Once the image diagnosis is established, the image can be used as a reference file for veterinary education and practice. There are many neuroradiology reference files or teaching files, either in printed or electronic forms, for human brain diseases (Poldrack and Gorgolewski 2014). However, few veterinary neuroradiology reference files are currently available. A few examples are case studies of intracranial arachnoid cysts in a chimpanzee (Kaneko et al. 2013; Miyabe-Nishiwaki et al. 2014) and an ependymal cyst in a cynomolgus macaque (Macaca fascicularis) (Bergin et al. 2008). We hope that our repository will provide educational files for veterinary neuroradiology.
Access to the Japan Monkey Centre Primates Brain Imaging Repository
This publication coincides with the public release of the JMC Primates Brain Imaging Repository. The images can be browsed online from the following JMC website (http://www.j-monkey.jp/BIR/index_e.html). Users can get access to the images and non-image information such as the cause of death and the autopsy findings after completion of a research agreement with the JMC collaborative research program.
Limitations
Our study had several limitations. First, the JMC’s brain samples were obtained from primates that had died of natural causes. Therefore, an accurate time of death was generally not known.
Second, in some primate postmortem brain samples, there are small incisions on the brain surface marked when the brain was extracted from the cranium. It is difficult to avoid making these small incisions when making postmortem brain samples.
Third, there was a brain size limitation. The bore diameter of the 9.4-T MRI equipment is approximately 200 mm, and the maximum diameter of the brain samples that can be scanned is approximately 70 mm. This size limitation is problematic in scanning brains of great apes having a size larger than 70 mm, such as brains of chimpanzees and gorillas. In the future, we plan to scan these brain samples of great apes using other scanners with larger bore diameter and open these data to the public.
Fourth, the users of this database should take account of the shrinkage of the brain size and the change of the brain morphology after the formalin fixation. The average shrinkage of the formalin-fixed brain is approximately 10–20% (Quester and Schröder 1997; Kinoshita et al. 2001; de Guzman et al. 2016). The degree of shrinkage also depends on the water content of the brain tissue. Especially, the degree of shrinkage of the non-myelinated immature brain samples, which contain water-rich tissue, would be greater than that of mature brain samples (Kinoshita et al. 2001). Our database includes immature brain samples such as the Toque macaque’s brain sample (Pr6182, 1 year of age after birth). Therefore, the shrinkage rates should be considered when analyzing the size and shape of the formalin-fixed MRIs of the brains, especially non-myelinated brains.
Fifth, there was an age variation in the animals whose brain samples were used to obtain MRI data. The animals ranged from 1 year of age (Toque macaque, Pr6182) to 29 years of age (Crab-eating Macaque, Pr6413) or “old” (Japanese Macaque, Pr6158; Japanese Macaque, Pr6378; Red-tailed Monkey, Pr6379) in this database. In this study, as a first step, we focused on how to create a new scan protocol for brain samples with various sizes using 9.4 T MRI. Because we selected the brain samples that had good preservation and few incisions on the brain surface for scans, there was an age variation in brain samples. Immature brains are still undergoing development. Aged brains have shrinkage of the brain size and expansion of the ventricles. Moreover, these brains contain amyloid plaques, which are known as a symptom of Alzheimer’s disease (Yamashita et al. 2012). As a next step, we will try to accumulate brain images of fully adult primates as far as possible to establish a standard brain image database.
Sixth, some brain images have image artifacts or insufficient SNR. These problems are caused by air bubbles from the brain sample and the container that severely decrease the quality of ex vivo MRI data. In order to solve the problem of air bubbles, we will try to completely eliminate air bubbles from both the brain sample and the container by deaeration with a vacuum pump before scanning.
Finally, brain images of large brain samples have concentricum contrast inhomogeneity. This phenomenon was not reported in the past. It is standard practice to immerse the fixed brain samples in GD-PBS for more than 1 week prior to scanning for ex vivo MRI scanning of mouse brain samples (Johnson et al. 2002). Although this preprocessing was also applied for the ex vivo MRI scanning of marmoset brain samples (Hikishima et al. 2013), we found that it cannot be applied for that of larger brain samples, such as those of macaques and gibbons. The concentric contrast inhomogeneity appears to be due to non-uniform penetration of GD-PBS into the brain samples, but it is difficult to explain the exact cause of this artifact because GD usually enhances image contrast. Determining the mechanism that produces this image artifact will require further investigation.
Conclusions
The JMC Primates Brain Imaging Repository is an ex vivo MRI database of non-human primate brains with high-spatial-resolution that can facilitate scientific discoveries in the field of comparative neuroscience and brain evolution. As an initial release, a digital collection of structural MRIs and DTIs obtained from 12 species was opened to the research community. The repository is also important for preserving information about the brains of various primates, including endangered species, in a permanent digital form, for use in optimizing the methods of scanning large fixed brains, and for veterinary neuroradiology education. We will continue to develop this repository to achieve our goal of collecting brain MRIs of more than 100 non-human primate species.
References
Barton R (2006) Primate brain evolution: integrating comparative, neurophysiological, and ethological data. Evol Anthropol Issues News Rev 15(6):224–236
Barton RA, Harvey PHJN (2000) Mosaic evolution of brain structure in mammals. Nature 405(6790):1055–1058
Basser PJ, Mattiello J, LeBihan D (1994a) Estimation of the effective self-diffusion tensor from the NMR spin echo. J Magn Reson Ser B 103(3):247–254
Basser PJ, Mattiello J, LeBihan D (1994b) MR diffusion tensor spectroscopy and imaging. Biophys J 66(1):259–267. https://doi.org/10.1016/S0006-3495(94)80775-1
Bergin IL, Campbell B, Agnew DW (2008) Ependymal cyst in a cynomolgus macaque (Macaca fascicularis). J Med Primatol 37(5):239–244
Bottomley PA, Foster TH, Argersinger RE, Pfeifer LM (1984) A review of normal tissue hydrogen NMR relaxation times and relaxation mechanisms from 1–100 MHz: dependence on tissue type, NMR frequency, temperature, species, excision, and age. Med Phys 11(4):425–448
Charvet CJ, Hof PR, Raghanti MA, Kouwe AJ, Sherwood CC, Takahashi E (2017) Combining diffusion magnetic resonance tractography with stereology highlights increased cross-cortical integration in primates. J Comp Neurol 525(5):1075–1093
D’Arceuil H, de Crespigny A (2007) The effects of brain tissue decomposition on diffusion tensor imaging and tractography. NeuroImage 36(1):64–68
De Guzman AE, Wong MD, Gleave JA, Nieman BJ (2016) Variations in post-perfusion immersion fixation and storage alter MRI measurements of mouse brain morphometry. NeuroImage 142:687–695
Dunbar R, Shultz S (2017) Why are there so many explanations for primate brain evolution? Phil Trans R Soc B 372(1727):20160244
Estrada A, Garber PA, Rylands AB, Roos C, Fernandez-Duque E, Di Fiore A, Nekaris KA-I, Nijman V, Heymann EW, Lambert JE (2017) Impending extinction crisis of the world’s primates: why primates matter. Sci Adv 3(1):e1600946
Fix M, Sieber M, Overs M, Schäfer HJ, Galla H-J (2000) Temperature-dependent phase behaviour of dihydroxy octadecanoic acid methyl esters: influence of stereochemistry and position of the second polar moiety. Phys Chem Chem Phys 2(20):4515–4520
Hikishima K, Sawada K, Murayama A, Komaki Y, Kawai K, Sato N, Inoue T, Itoh T, Momoshima S, Iriki A (2013) Atlas of the developing brain of the marmoset monkey constructed using magnetic resonance histology. Neuroscience 230:102–113
Hofman M, Falk D (2012) Hominin paleoneurology: where are we now? Evolution of the primate brain: from neuron to behavior., pp 195–255
Holloway RL, Clarke RJ, Tobias PV (2004) Posterior lunate sulcus in Australopithecus africanus: was Dart right? C R Palevol 3(4):287–293
Jerison H (1973) Evolution of the brain and intelligence. Academic Press, Cambridge, New york and London
Jiang H, van Zijl PC, Kim J, Pearlson GD, Mori S (2006) DtiStudio: resource program for diffusion tensor computation and fiber bundle tracking. Comput Methods Progr Biomed 81(2):106–116. https://doi.org/10.1016/j.cmpb.2005.08.004
Johnson GA, Cofer GP, Gewalt SL, Hedlund LW (2002) Morphologic phenotyping with MR microscopy: the visible mouse. Radiology 222(3):789–793. https://doi.org/10.1148/radiol.2223010531
Kaas JH (2017) The evolution of mammalian brains from early mammals to present-day primates. Evolution of the brain, cognition, and emotion in vertebrates. Springer, Berlin, pp 59–80
Kaneko T, Sakai T, Miyabe-Nishiwaki T, Tomonaga M (2013) A case of naturally occurring visual field loss in a chimpanzee with an arachnoid cyst. Neuropsychologia 51(13):2856–2862
Kinoshita Y, Okudera T, Tsuru E, Yokota A (2001) Volumetric analysis of the germinal matrix and lateral ventricles performed using MR images of postmortem fetuses. Am J Neuroradiol 22(2):382–388
Kroenke CD, Bretthorst GL, Inder TE, Neil JJ (2005) Diffusion MR imaging characteristics of the developing primate brain. NeuroImage 25(4):1205–1213. https://doi.org/10.1016/j.neuroimage.2004.12.045
Krubitzer L, Kaas J (2005) The evolution of the neocortex in mammals: how is phenotypic diversity generated? Curr Opin Neurobiol 15(4):444–453
Makris N, Worth A, Papadimitriou G, Stakes J, Caviness V, Kennedy D, Pandya D, Kaplan E, Sorensen A, Wu O (1997) Morphometry of in vivo human white matter association pathways with diffusion-weighted magnetic resonance imaging. Ann Neurol 42(6):951–962
Markov NT, Ercsey-Ravasz M, Ribeiro Gomes A, Lamy C, Magrou L, Vezoli J, Misery P, Falchier A, Quilodran R, Gariel M (2012) A weighted and directed interareal connectivity matrix for macaque cerebral cortex. Cereb Cortex 24(1):17–36
Mars RB, Neubert F-X, Verhagen L, Sallet J, Miller KL, Dunbar RI, Barton RA (2014) Primate comparative neuroscience using magnetic resonance imaging: promises and challenges. Front Neurosci 8:298
Miyabe-Nishiwaki T, Kaneko T, Sakai T, Kaneko A, Watanabe A, Watanabe S, Maeda N, Kumazaki K, Suzuki J, Fujiwara R (2014) Intracranial arachnoid cysts in a chimpanzee (Pan troglodytes). Primates 55(1):7–12
Morecraft RJ, Ugolini G, Lanciego JL, Wouterlood FG, Pandya DN (2009) Classic and contemporary neural tract tracing techniques. Diffusion MRI: from quantitative measurement to in vivo neuroanatomy. Elsevier, Amsterdam, pp 273–308
Mori S, van Zijl P (2002) Fiber tracking: principles and strategies—a technical review. NMR Biomed 15(7–8):468–480
Mori S, Zhang J (2006) Principles of diffusion tensor imaging and its applications to basic neuroscience research. Neuron 51(5):527–539
Mori S, Wakana S, Van Zijl PC, Nagae-Poetscher L (2005) MRI atlas of human white matter. Elsevier, Netherland
Oishi K, Huang H, Yoshioka T, Ying SH, Zee DS, Zilles K, Amunts K, Woods R, Toga AW, Pike GB (2011) Superficially located white matter structures commonly seen in the human and the macaque brain with diffusion tensor imaging. Brain Connect 1(1):37–47
Okano H, Nomura T (2008) Brain MR atlas of common marmoset (in Japanese). Yodosha, Tokyo
Pajevic S, Pierpaoli C (1999) Color schemes to represent the orientation of anisotropic tissues from diffusion tensor data: application to white matter fiber tract mapping in the human brain. Magn Reson Med 42(3):526–540
Paxinos G, Huang X-F, Toga AW (2000) The rhesus monkey brain in stereotaxic coordinates. Academic Press, San Diego
Pierpaoli C, Barnett A, Pajevic S, Chen R, Penix L, Virta A, Basser P (2001) Water diffusion changes in Wallerian degeneration and their dependence on white matter architecture. NeuroImage 13(6):1174–1185
Poldrack RA, Gorgolewski KJ (2014) Making big data open: data sharing in neuroimaging. Nat Neurosci 17(11):1510–1517
Preuss TM (2011) The human brain: rewired and running hot. Ann N Y Acad Sci 1225(Suppl 1):E182–191. https://doi.org/10.1111/j.1749-6632.2011.06001.x
Quester R, Schröder R (1997) The shrinkage of the human brain stem during formalin fixation and embedding in paraffin. J Neurosci Methods 75(1):81–89
Rilling JK (2014) Comparative primate neuroimaging: insights into human brain evolution. Trends Cognit Sci 18(1):46–55
Rilling JK, Glasser MF, Preuss TM, Ma X, Zhao T, Hu X, Behrens TE (2008) The evolution of the arcuate fasciculus revealed with comparative DTI. Nat Neurosci 11(4):426
Roth G, Dicke U (2005) Evolution of the brain and intelligence. Trends Cognit Sci 9(5):250–257
Saleem KS, Logothetis NK (2012) A combined MRI and histology atlas of the rhesus monkey brain in stereotaxic coordinates. Academic Press, Netherlands
Sawada K, Sun X-Z, Fukunishi K, Kashima M, Sakata-Haga H, Tokado H, Aoki I, Fukui Y (2009) Developments of sulcal pattern and subcortical structures of the forebrain in cynomolgus monkey fetuses: 7-Tesla magnetic resonance imaging provides high reproducibility of gross structural changes. Brain Struct Funct 213(4–5):469–480
Sawada K, Fukunishi K, Kashima M, Saito S, Sakata-Haga H, Aoki I, Fukui Y (2012) Fetal gyrification in cynomolgus monkeys: a concept of developmental stages of gyrification. Anat Rec 295(7):1065–1074
Sawada K, Hikishima K, Murayama A, Okano H, Sasaki E, Okano H (2014) Fetal sulcation and gyrification in common marmosets (Callithrix jacchus) obtained by ex vivo magnetic resonance imaging. Neuroscience 257:158–174
Schmahmann JD, Pandya DN, Wang R, Dai G, D’arceuil HE, de Crespigny AJ, Wedeen VJ (2007) Association fibre pathways of the brain: parallel observations from diffusion spectrum imaging and autoradiography. Brain 130(3):630–653
Shatil AS, Matsuda KM, Figley CR (2016) A method for whole brain ex vivo magnetic resonance imaging with minimal susceptibility artifacts. Front Neurol 7:208
Shepherd TM, Flint JJ, Thelwall PE, Stanisz GJ, Mareci TH, Yachnis AT, Blackband SJ (2009) Postmortem interval alters the water relaxation and diffusion properties of rat nervous tissue—implications for MRI studies of human autopsy samples. NeuroImage 44(3):820–826
Woods RP, Grafton ST, Holmes CJ, Cherry SR, Mazziotta JC (1998) Automated image registration: I. General methods and intrasubject, intramodality validation. J Comput Assisted Tomogr 22(1):139–152
Yamashita A, Fuchs E, Taira M, Yamamoto T, Hayashi M (2012) Somatostatin-immunoreactive senile plaque-like structures in the frontal cortex and nucleus accumbens of aged tree shrews and Japanese macaques. J Med Primatol 41(3):147–157
Zhang K, Sejnowski TJ (2000) A universal scaling law between gray matter and white matter of cerebral cortex. Proc Nat Acad Sci 97(10):5621–5626
Acknowledgements
We thank Drs. R. Sakamoto and D. Wu for technical advice on imaging analysis, Dr. Y. Tomogane for image diagnosis. We also thank Profs. A. Mikami and H. Imai for advice about primate brain samples, and Profs. T. Matsuzawa, M. Tomonaga, and J. Yamagiwa for permitting us to use the JMC primate brain sample collection. We thank Dr. E. Nakajima for help with manuscript editing. This work was financially supported by JSPS KAKENHI Grant for Young Scientists (B) (#26870827 and # 17K18097 to T.S.), a JSPS Postdoctoral Fellowship for Research Abroad (#490 to T.S.), Brain/MINDS by AMED (JP17dm0207002 to T.S./S.S., JP17dm0207001 to H.O./J.H.), and the Fakhri Rad Brite Star award from the Department of Radiology Johns Hopkins University School of Medicine (to K.O.). This work was also supported by the JMC collaborative research program (#2014013, #2015019, and #2016017) and the Cooperative Research Program of the Primate Research Institute, Kyoto University (#H25-E31 and# H26-C7 to H.O, and #H27-D23 to T.S.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Sakai, T., Hata, J., Ohta, H. et al. The Japan Monkey Centre Primates Brain Imaging Repository for comparative neuroscience: an archive of digital records including records for endangered species. Primates 59, 553–570 (2018). https://doi.org/10.1007/s10329-018-0694-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10329-018-0694-3