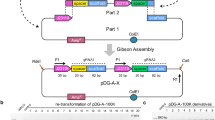
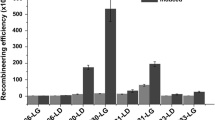
Abstract
Although CRISPR/Cas9-mediated gene editing technology has developed vastly in Escherichia coli, the chromosomal integration of large DNA fragment is still challenging compared with gene deletion and small fragment integration. Moreover, to guarantee sufficient Cas9-induced double-strand breaks, it is usually necessary to design several gRNAs to select the appropriate one. Accordingly, we established a practical daily routine in the laboratory work, involving multiple-step chromosomal integration of the divided segments from a large DNA fragment. First, we introduced and optimized the protospacers from Streptococcus pyogenes in E. coli W3110. Next, the appropriate fragment size for each round of integration was optimized to be within 3–4 kb. Taking advantage of the optimized protospacer/gRNA pairs, a DNA fragment with a total size of 15.4 kb, containing several key genes for uridine biosynthesis, was integrated into W3110 chromosome, which produced 5.6 g/L uridine in shake flask fermentation. Using this strategy, DNA fragments of virtually any length can be integrated into a suitable genomic site, and two gRNAs can be alternatively used, avoiding the tedious construction of gRNA-expressing plasmids. This study thus presents a useful strategy for large DNA fragment integration into the E. coli chromosome, which can be easily adapted for use in other bacteria.




Similar content being viewed by others
References
Atsumi S, Hanai T, Liao JC (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86–89. https://doi.org/10.1038/nature06450
Cunningham DS, Koepsel RR, Ataai MM, Domach MM (2009) Factors affecting plasmid production in Escherichia coli from a resource allocation standpoint. Microb Cell Fact 8:27. https://doi.org/10.1186/1475-2859-8-27
Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E (2011) CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471:602–607. https://doi.org/10.1038/nature09886
Esvelt KM, Wang HH (2013) Genome-scale engineering for systems and synthetic biology. Mol Syst Biol 9:641. https://doi.org/10.1038/msb.2012.66
Fan XG, Wu HY, Li GL, Yuan H, Zhang HC, Li YJ, Xie XX, Chen N (2017) Improvement of uridine production of Bacillus subtilis by atmospheric and room temperature plasma mutagenesis and high-throughput screening. PLoS One 12:e0176545. https://doi.org/10.1371/journal.pone.0176545
Garneau JE, Dupuis MÈ, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S (2010) The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67. https://doi.org/10.1038/nature09523
Jeong J, Cho N, Jung D, Bang D (2013) Genome-scale genetic engineering in Escherichia coli. Biotechnol Adv 31:804–810. https://doi.org/10.1016/j.biotechadv.2013.04.003
Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA (2013) RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31:233–239. https://doi.org/10.1038/nbt.2508
Jiang Y, Chen B, Duan C, Sun B, Yang J, Yang S (2015) Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol 81:2506–2514. https://doi.org/10.1128/AEM.04023-14
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. https://doi.org/10.1126/science.1225829
Kuhlman TE, Cox EC (2010) Site-specific chromosomal integration of large synthetic constructs. Nucleic Acids Res 38:e92. https://doi.org/10.1093/nar/gkp1193
Lange J, Müller F, Takors R, Blombach B (2018) Harnessing novel chromosomal integration loci to utilize an organosolv-derived hemicellulose fraction for isobutanol production with engineered Corynebacterium glutamicum. Microb Biotechnol 11:257–263. https://doi.org/10.1111/1751-7915.12879
Li YF, Lin ZQ, Huang C, Zhang Y, Wang Z, Tang YJ (2015) Metabolic engineering of Escherichia coli using CRISPR-Cas9 meditated genome editing. Metab Eng 31:13–21. https://doi.org/10.1016/j.ymben.2015.06.006
Nakashima N, Akita H, Hoshino T (2014) Establishment of a novel gene expression method, BICES (biomass-inducible chromosome-based expression system), and its application to the production of 2,3-butanediol and acetoin. Metab Eng 25:204–214. https://doi.org/10.1016/j.ymben.2014.07.011
Park HM, Liu H, Wu J, Chong A, Mackley V, Fellmann C, Rao A, Jiang F, Chu H, Murthy N, Lee K (2018) Extension of the crRNA enhances Cpf1 gene editing in vitro and in vivo. Nat Commun 9:3313. https://doi.org/10.1038/s41467-018-05641-3
Pyne ME, Mooyoung M, Chung DA, Chou CP (2015) Coupling the CRISPR/Cas9 system with lambda red recombineering enables simplified chromosomal gene replacement in Escherichia coli. Appl Environ Microbiol 81:5103–5114. https://doi.org/10.1128/AEM.01248-15
Sharan SK, Thomason LC, Kuznetsov SG, Court DL (2009) Recombineering: a homologous recombination-based method of genetic engineering. Nat Protoc 4:206. https://doi.org/10.1038/nprot.2008.227
Song CW, Lee J, Lee SY (2015) Genome engineering and gene expression control for bacterial strain development. Biotechnol J 10:56–68. https://doi.org/10.1002/biot.201400057
St-Pierre F, Cui L, Priest DG, Endy D, Dodd IB, Shearwin KE (2013) One-step cloning and chromosomal integration of DNA. ACS Synth Biol 2:537–541. https://doi.org/10.1021/sb400021j
Tyo KEJ, Ajikumar PK, Stephanopoulos G (2009) Stabilized gene duplication enables long-term selection-free heterologous pathway expression. Nat Biotechnol 27:760–765. https://doi.org/10.1038/nbt.1555
Ublinskaya AA, Samsonov VV, Mashko SV, Stoynova NV (2012) A PCR-free cloning method for the targeted φ80 Int-mediated integration of any long DNA fragment, bracketed with meganuclease recognition sites, into the Escherichia coli chromosome. J Microbiol Methods 89:167–173. https://doi.org/10.1016/j.mimet.2012.03.013
Wang HH, Church GM (2011) Multiplexed genome engineering and genotyping methods applications for synthetic biology and metabolic engineering. Method Enzymol 498:409–426. https://doi.org/10.1016/B978-0-12-385120-8.00018-8
Yadav VG, De Mey M, Lim CG, Ajikumar PK, Stephanopoulos G (2012) The future of metabolic engineering and synthetic biology: towards a systematic practice. Metab Eng 14:233–241. https://doi.org/10.1016/j.ymben.2012.02.001
Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL (2000) An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA 97:5978–5983. https://doi.org/10.1073/pnas.100127597
Acknowledgements
We would like to thank Prof. Tao Chen of Tianjin University for providing plasmids used in the CRISPR/Cas9 system. This work was financially supported by National High Technology Research and Development Program (2015AA021003), National Natural Science Foundation of China (31500026, 31700037), China Postdoctoral Science Foundation funded project (2016M601269, 2017M61170) and Tianjin Key Technology R & D program of Tianjin Municipal Science and Technology Commission (17YFZCSY01050).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Y., Yan, F., Wu, H. et al. Multiple-step chromosomal integration of divided segments from a large DNA fragment via CRISPR/Cas9 in Escherichia coli. J Ind Microbiol Biotechnol 46, 81–90 (2019). https://doi.org/10.1007/s10295-018-2114-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-018-2114-5