Abstract
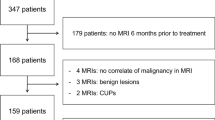
We quantitatively investigate the influence of image registration, using open-source software (3DSlicer), on kinetic analysis (Tofts model) of dynamic contrast enhanced MRI of early-stage breast cancer patients. We also show that registration computation time can be reduced by reducing the percent sampling (PS) of voxels used for estimation of the cost function. DCE-MRI breast images were acquired on a 3T-PET/MRI system in 13 patients with early-stage breast cancer who were scanned in a prone radiotherapy position. Images were registered using a BSpline transformation with a 2 cm isotropic grid at 100, 20, 5, 1, and 0.5PS (BRAINSFit in 3DSlicer). Signal enhancement curves were analyzed voxel-by-voxel using the Tofts kinetic model. Comparing unregistered with registered groups, we found a significant change in the 90th percentile of the voxel-wise distribution of Ktrans. We also found a significant reduction in the following: (1) in the standard error (uncertainty) of the parameter value estimation, (2) the number of voxel fits providing unphysical values for the extracellular-extravascular volume fraction (ve > 1), and (3) goodness of fit. We found no significant differences in the median of parameter value distributions (Ktrans, ve) between unregistered and registered images. Differences between parameters and uncertainties obtained using 100PS versus 20PS were small and statistically insignificant. As such, computation time can be reduced by a factor of 2, on average, by using 20PS while not affecting the kinetic fit. The methods outlined here are important for studies including a large number of post-contrast images or number of patient images.







Similar content being viewed by others
References
Khalifa F, Soliman A, El-Baz A, Abou El-Ghar M, El-Diasty T, Gimel’farb G, et al: Models and methods for analyzing DCE-MRI: A review. Med Phys 41:124301,2014. https://doi.org/10.1118/1.4898202
Wang C-H: Review of treatment assessment using DCE-MRI in breast cancer radiation therapy. World J Methodol 4:46,2014. https://doi.org/10.5662/wjm.v4.i2.46
O’connor J, Jackson A, Parker G, Jayson GC: DCE-MRI biomarkers in the clinical evaluation of antiangiogenic and vascular disrupting agents ANGIOGENESIS: A TARGET FOR ANTICANCER THERAPY. Br J Cancer 96:189–95,2007. https://doi.org/10.1038/sj.bjc.6603515
Giger ML, Karssemeijer N, Schnabel JA. Breast Image Analysis for Risk Assessment, Detection, Diagnosis, and Treatment of Cancer. Annu Rev Biomed Eng 15:327–57,2013. https://doi.org/10.1146/annurev-bioeng-071812-152416
Dmitriev ID, Loo CE, Vogel W V, Pengel KE, Gilhuijs AKG: Fully automated deformable registration of breast DCE-MRI and PET/CT. Phys Med Biol 58:1221–33,2013. https://doi.org/10.1088/0031-9155/58/4/1221
Denton ER, Sonoda LI, Rueckert D, Rankin SC, Hayes C, Leach MO, et al: Comparison and evaluation of rigid, affine, and nonrigid registration of breast MR images. J Comput Assist Tomogr 23:800–5,1999. https://doi.org/10.1097/00004728-199909000-00031
Hill A, Mehnert A, Crozier S, McMahon K: Evaluating the accuracy and impact of registration in dynamic contrast-enhanced breast MRI. Concepts Magn Reson Part B Magn Reson Eng 35:106–20,2009. https://doi.org/10.1002/cmr.b.20133
Rueckert D, Sonodaa LI, Denton E, Rankina S, Hayesc C, Leachc M, et al: Comparison and evaluation of rigid and non-rigid registration of breast MR images. Med Imaging 99:3661,1999, p. 78–88
Schäfer S, Preim U, Glaßer S, Preim B, Tönnies K: Local similarity measures for lesion registration in DCE - MRI of the breast. Ann BMVA 2011:1–13,2011
Martel AL, Froh MS, Brock KK, Plewes DB, Barber DC: Evaluating an optical-flow-based registration algorithm for contrast-enhanced magnetic resonance imaging of the breast. Phys Med Biol 52:3803–16,2007. https://doi.org/10.1088/0031-9155/52/13/010.
Woolf DK, Taylor NJ, Makris A, Tunariu N, Collins DJ, Li SP, et al: Arterial input functions in dynamic contrast-enhanced magnetic resonance imaging: which model performs best when assessing breast cancer response? Br J Radiol 89:20150961,2016. https://doi.org/10.1259/bjr.20150961
Rueckert D, Sonoda LI, Hayes C, Hill DL, Leach MO, Hawkes DJ: Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging 18:712–21,1999. https://doi.org/10.1109/42.796284
Tanner C, Schnabel JA., Hill DLG, Hawkes DJ, Degenhard A, Leach MO, et al: Quantitative evaluation of free-form deformation registration for dynamic contrast-enhanced MR mammography. Med Phys 34:1221,2007. https://doi.org/10.1118/1.2712040
Chebrolu VV, Shanbhag D, Bedair R, Gupta S, Hervo P, Reid S, et al: Impact of non-rigid motion correction on pharmaco-kinetic analysis for breast dynamic contrast-enhanced MRI. Proc 23rd Sci Meet Int Soc Magn Reson Med 2015:668
Guidolin K, Lock M, Yaremko B, Gelman N, Gaede S, Kornecki A, et al: A phase II trial to evaluate single-dose stereotactic body radiation therapy (SBRT) prior to surgery for early-stage breast carcinoma: SIGNAL (stereotactic image-guided neoadjuvant ablative radiation then lumpectomy) trial. J Radiat Oncol 4:423–30,2015. https://doi.org/10.1007/s13566-015-0227-2.
Johnson HJ, Mccormick MM, Ibanez L: The ITK Software Guide Book 1: Introduction and Development Guidelines Fourth Edition Updated for ITK version 4. 8 2015. https://itk.org/
Parker GJM, Roberts C, Macdonald A, Buonaccorsi GA, Cheung S, Buckley DL, et al: Experimentally-derived functional form for a population-averaged high-temporal-resolution arterial input function for dynamic contrast-enhanced MRI. Magn Reson Med 56:993–1000,2006. https://doi.org/10.1002/mrm.21066
Patrick JC, Terry Thompson R, So A, Butler J, Faul D, Stodilka RZ, et al: Technical Note: Comparison of megavoltage, dual-energy, and single-energy CT-based μ-maps for a four-channel breast coil in PET/MRI. Med Phys 44:4758–65,2017. https://doi.org/10.1002/mp.12407
Johnson HJ, Harris G, Williams K, Williams NK, Williams K: BRAINSFit: Mutual Information Rigid Registrations of Whole-Brain 3D Images, Using the Insight Toolkit. 2007.
Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin J-C, Pujol S, et al: 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 30:1323–41,2012. https://doi.org/10.1016/j.mri.2012.05.001
Kikinis R, Pieper SD, Vosburgh KG: 3D Slicer: A Platform for Subject-Specific Image Analysis, Visualization, and Clinical Support. Intraoperative Imaging Image-Guided Ther 2014. https://doi.org/10.1007/978-1-4614-7657-3_19
Kapur T, Pieper S, Fedorov A, Fillion-Robin JC, Halle M, O’Donnell L, et al. Increasing the impact of medical image computing using community-based open-access hackathons: The NA-MIC and 3D Slicer experience. Med Image Anal 2016. https://doi.org/10.1016/j.media.2016.06.035
Melbourne A, Hipwell J, Modat M, Mertzanidou T, Huisman H, Ourselin S, et al: The effect of motion correction on pharmacokinetic parameter estimation in dynamic-contrast-enhanced MRI. Phys Med Biol 56:7693–708,2011. https://doi.org/10.1088/0031-9155/56/24/001
Otsu N: A Threshold Selection Method from Gray-Level Histograms. IEEE Trans Syst Man Cybern 9:62–6,1979. https://doi.org/10.1109/TSMC.1979.4310076
Tofts P, Gunnar B, Buckley DL, Evelhoch JL, Henderson E, Knopp M V, et al: Estimating kinetic parameters from dynamic contrast-enhanced T1-weighted MRI of a diffusable tracer: Standardized quantities and symbols. J Magn Reson Imaging 10:223–32,1999
Tofts PS, Parker GJM: DCE-MRI: Acquisition and analysis techniques. In: Barker P, Golay X, Zaharchuk G, Eds. Clin. Perfus. MRI Tech. Appl., vol. 9781107013, Cambridge: Cambridge University Press, 2010, pp 58–74. https://doi.org/10.1017/CBO9781139004053.006
Hayes C, Padhani AR, Leach MO: Assessing changes in tumour vascular function using dynamic contrast-enhanced magnetic resonance imaging. NMR Biomed 15:154–63,2002. https://doi.org/10.1002/nbm.756
Yankeelov TE, Lepage M, Chakravarthy A, Broome EE, Niermann KJ, Kelley MC, et al: Integration of quantitative DCE-MRI and ADC mapping to monitor treatment response in human breast cancer : initial results. Magn Reson Imaging 25:1–13,2007. https://doi.org/10.1016/j.mri.2006.09.006
Acknowledgments
The authors would like to thank Siemens Canada for supplying the breast coil used in this study.
Funding
The authors would also like to acknowledge grant funding from the Ontario Research Fund–Research Excellence (OCAIRO Project RE-04-026), the Canadian institutes of Health Research (149080), and the Breast Cancer Society of Canada for funding support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mouawad, M., Biernaski, H., Brackstone, M. et al. The Effect of Registration on Voxel-Wise Tofts Model Parameters and Uncertainties from DCE-MRI of Early-Stage Breast Cancer Patients Using 3DSlicer. J Digit Imaging 33, 1065–1072 (2020). https://doi.org/10.1007/s10278-020-00374-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-020-00374-6