Abstract
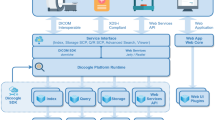
Especially for investigator-initiated research at universities and academic institutions, Internet-based rare disease registries (RDR) are required that integrate electronic data capture (EDC) with automatic image analysis or manual image annotation. We propose a modular framework merging alpha-numerical and binary data capture. In concordance with the Office of Rare Diseases Research recommendations, a requirement analysis was performed based on several RDR databases currently hosted at Uniklinik RWTH Aachen, Germany. With respect to the study management tool that is already successfully operating at the Clinical Trial Center Aachen, the Google Web Toolkit was chosen with Hibernate and Gilead connecting a MySQL database management system. Image and signal data integration and processing is supported by Apache Commons FileUpload-Library and ImageJ-based Java code, respectively. As a proof of concept, the framework is instantiated to the German Calciphylaxis Registry. The framework is composed of five mandatory core modules: (1) Data Core, (2) EDC, (3) Access Control, (4) Audit Trail, and (5) Terminology as well as six optional modules: (6) Binary Large Object (BLOB), (7) BLOB Analysis, (8) Standard Operation Procedure, (9) Communication, (10) Pseudonymization, and (11) Biorepository. Modules 1–7 are implemented in the German Calciphylaxis Registry. The proposed RDR framework is easily instantiated and directly integrates image management and analysis. As open source software, it may assist improved data collection and analysis of rare diseases in near future.





Similar content being viewed by others
Notes
References
Rubinstein YR, Groft SC, Bartek R, Brown K, Christensen RA, Collier E, Farber A, Farmer J, Ferguson JH, Forrest CB, Lockhart NC, McCurdy KR, Moore H, Pollen GB, Rangel Miller RV, Hullm S, Vaught J: Creating a global rare disease patient registry linked to a rare diseases biorepository database: Rare Disease-HUB (RD-HUB). Contemp Clin Trials 31:394–404, 2010
Bellgard M, Beroud C, Parkinson K, Harris T, Ayme S, Baynam G, Weeramanthri T, Dawkins H, Hunter A: Dispelling myths about rare disease registry system development. Source Code Biol Med 8(1):21, 2013
Bellgard MI, Macgregor A, Janon F, Harvey A, O’Leary P, Hunter A, Dawkins H: A modular approach to disease registry design: successful adoption of an internet-based rare disease registry. Hum Mutat 33(10):E2356–E2366, 2012
Messiaen C, Le Mignot L, Rath A, Richard JB, Dufour E, Ben Said M, Jais JP, Verloes A, Le Merrer M, Bodemer C, Baujat G, Gerard-Blanluet M, Bourdon-Lanoy E, Salomon R, Ayme S, Landais P: CEMARA: a Web dynamic application within a N-tier architecture for rare diseases. Stud Health Technol Inform 136:51–56, 2008
Seo H, Kim D, Chae JH, Kang HG, Lim BC, Cheong HI, Kim JH: Development of Korean rare disease knowledge base. Healthc Inform Res 18(4):272–278, 2012
Stausberg J, Altmann A, Antony G, Drepper J, Sax U, Schütt A: Register for networked medical research in Germany. Appl Clin Inform 1(4):408–418, 2010
Drepper J, Semler SC (ed). IT infrastructure in patient-centered research. State of the art and required actions (German). Technical report. IT-Reviewing Board of TMF. Akademische Verlagsgesellschaft AKA GmbH, Berlin, 2013 (ISBN 978-3-89838-690-6)
Ayme S (ed). Disease Registries in Europe. Orphanet Report Series, Rare Diseases Collection. Orphanet Technical Report. Jan 2013. Availabe at: http://www.orpha.net/orphacom/cahiers/docs/GB/Registries.pdf
Souza MP, Miller VR: Significance of patient registries for dermatological disorders. J Investig Dermatol 132:1749–1752, 2012
Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG: Research electronic data capture (REDCap) – A metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 42(2):377–381, 2009
Natter MD, Quan J, Ortiz DM, Bousvaros A, Ilowite NT, Inman CJ, Marsolo K, McMurry AJ, Sandborg CI, Schanberg LE, Wallace CA, Warren RW, Weber GM, Mandl KD: An i2b2-based, generalizable, open source, self-scaling chronic disease registry. J Am Med Inform Assoc 20:172–179, 2013
Kohane IS, Churchill SE, Murphy SN: A translational engine at the national scale: informatics for integrating biology and the bedside. J Am Med Inform Assoc 19:181e5, 2012
Wang X, Martinez C, Wang J, Liu Y, Liu BJ. Development of a user customizable imaging informatics-based intelligent workflow engine system to enhance rehabilitation clinical trials. Proceedings SPIE 2014; 9039, in press
Langer S, Bartholmai B: Imaging informatics: challenges in multi-site imaging trials. J Digit Imaging 24(1):151–159, 2011. doi:10.1007/s10278-010-9282-9
Erickson BJ, Pan T, Marcus DS: CTSA Imaging Informatics Project Group. Whitepapers on imaging infrastructure for research: Part 1: General workflow considerations. J Digit Imaging 25(4):449–453, 2012
Marcus DS, Erickson BJ, Pan T: Imaging infrastructure for research. Part 2. Data management practices. J Digit Imaging 25(5):566–569, 2012
Prins AH, Abu-Hanna A: Requirements analysis of information services for patients on a general practitioner's website–patient and general practitioner's perspectives. Methods Inf Med 46(6):629–635, 2007
Prokosch HU, Beck A, Ganslandt T, Hummel M, Kiehntopf M, Sax U, Ückert F, Semler S: IT Infrastructure Components for Biobanking. Appl Clin Inform 1(4):419–429, 2010
Deserno TM, Deserno V, Legewie V, Schafhausen J, Eisert A, Schmidt-Kotsas A, Kirstein S, Willems J, Spitzer K, Schulz JB. IT support for translational management of clinical trials based on the Google Web Toolkits. German Medical Science (GMS) Meeting Abstract 2011: 11gmds023. (German)
Brandenburg VM, Kramann R, Specht P, Ketteler M: Calciphylaxis in CKD and beyond. Nephrol Dial Transplant 27(4):1314–1318, 2012
Brandenburg V, Specht P, Floege J, Ketteler M. Seven Years of Experience with the German CUA Registry. Abstract Presentation. American Society of Nephrology Renal Week, 2013
Deserno TM, Sarandi I, Jose A, Haak D, Jonas S, Specht P, Brandenburg V. Towards quantitative Calciphylaxis. Proceedings SPIE 2014; 9035: in press
Jäkel J, Schmidt R, Leusmann P, Spreckelsen C, Knüchel R, Dahl E, Veeck J: Biospecimen quality management at the RWTH centralized biomaterial bank (RWTH cBMB). Pathologe 34(1):136, 2013
Hirschberg I, Knüppel H, Strech D: Practice variation across consent templates for biobank research. a survey of German biobanks. Front Genet 4:240, 2013
Nebel E, Masinter L. Form-based file upload in HTML. Request for Comments no. 1867. The Internet Engineering Task Force (IETF), Network Working Group. 1995. Available at http://www.ietf.org/rfc/rfc1867.txt
Deserno TM, Haak D, Samsel C, Gehlen J, Kabino K. Integration image management and analysis into OpenClinica using web services. Proc SPIE 8674: 0F1-10, 2013
Wagner M, Glock J, Sariyar M, Borg A. PID Generator. Technical Manual. Version 1.2. Dept. of Medical Informatics and Biometry. Johannes-Gutenberg-Universität Mainz, Germany, 2008
Lehmann TM, Schubert H, Keysers D, Kohnen M, Wein BB: The IRMA code for unique classification of medical images. Proc SPIE 5033:440–451, 2003
Digital Imaging and Communications in Medicine (DICOM). Part 19: Application Hosting. PS3.19-2011. National Electrical Manufacturers Association, Rosslyn, VA, USA, 2011
Prior FW, Erickson BJ, Tarbox L: Open source software projects of the caBIG In Vivo Imaging Workspace Software special interest group. J Digit Imaging 20(Suppl 1):94–100, 2007
Channin DS, Mongkolwat P, Kleper V, Sepukar K, Rubin DL: The caBIG annotation and image markup project. J Digit Imaging 23(2):217–225, 2010
Ohmann C, Kuchinke W: Future developments of medical informatics from the viewpoint of networked clinical research. Interoperability and integration. Methods Inf Med 48(1):45–54, 2009
Fridsma DB, Evans J, Hastak S, Mead CN: The BRIDG project: a technical report. J Am Med Inform Assoc 15(2):130–137, 2008
Hullsiek KH, George M, Brown SK: Designing and managing a flexible and dynamic biorepository system: a 15-year perspective from the CPCRA, ESPRIT, and INSIGHT clinical trial networks. Curr Opin HIV AIDS 5(6):538–544, 2010
Lin MC, Vreeman DJ, McDonald CJ, Huff SM: A characterization of local LOINC mapping for laboratory tests in three large institutions. Methods Inf Med 50(2):105–114, 2011
Dugas M, Thun S, Frankewitsch T, Heitmann KU: LOINC codes for hospital information systems documents: a case study. J Am Med Inform Assoc 16(3):400–403, 2009
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Deserno, T.M., Haak, D., Brandenburg, V. et al. Integrated Image Data and Medical Record Management for Rare Disease Registries. A General Framework and its Instantiation to the German Calciphylaxis Registry. J Digit Imaging 27, 702–713 (2014). https://doi.org/10.1007/s10278-014-9698-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-014-9698-8