Abstract
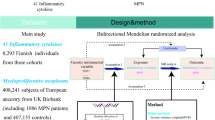
Diffuse large B cell lymphoma (DLBCL) is the most common type of non-Hodgkin lymphoma. Studies indicated that inflammatory cytokines involved in the occurrence and progression of DLBCL and it is challenging to discern causality from the effects due to the presence of feedback loops. We conducted a bidirectional Mendelian randomization (MR) study to investigate the potential causal relationship between DLBCL and inflammatory cytokines. The genetic variants associated with inflammatory cytokines were obtained from a genome-wide association study (GWAS) involving 8293 European participants, and the data on 1010 individuals with DLBCL were sourced from the FinnGen consortium. The primary method employed in this study was the inverse-variance weighted (IVW) method, with supplementary analyses conducted using the MR-Egger, weighted median, and MR-PRESSO approaches. Based on the IVW method, genetically predicted that increasing level of Monokine induced by interferon gamma (MIG/CXC chemokine ligand 9, CXCL9) [OR: 1.31; 95% CI: 1.05–1.62; P = 0.01] and interferon gamma-induced protein 10(IP-10/CXC chemokine ligand 10, CXCL10) [OR: 1.30; 95% CI: 1.02–1.66; P = 0.03] showed suggestive associations with DLBCL risk. DLBCL may increase the level of macrophage colony-stimulating factor (M-CSF) [OR: 1.12; 95% CI: 1.01–1.2; P = 0.03], tumor necrosis factor beta (TNF-β) [OR: 1.16; 95% CI: 1.02–1.31; P = 0.02] and TNF-related apoptosis-inducing ligand (TRAIL) [OR: 1.07; 95% CI: 1.01–1.13; P = 0.02]. This study presents evidence supporting a causal relationship between inflammation cytokines and DLBCL. Specifically, MIG/CXCL9 and IP-10/CXCL10 were identified as indicators of upstream causes of DLBCL; while, DLBCL itself was found to elevate the levels of M-CSF, TNF-β, and TRAIL. These findings suggest that targeting specific inflammatory factors through regulation and intervention could serve as a potential approach for the treatment and prevention of DLBCL.





Similar content being viewed by others
Data availability
We thank the participants and researchers for providing the publicly available summary data used in this study. The data sources and handling of these data are described in the Materials and Methods and in the Supplementary Tables. Further information is available from the corresponding author upon request.
Abbreviations
- DLBCL:
-
Diffuse large B cell lymphoma
- MR:
-
Mendelian randomization
- IVs:
-
Instrumental variables
- GWASs:
-
Genome-wide association studies
- CI:
-
Confidence interval
- IVW:
-
Inverse-variance weighted
- MR-PRESSO:
-
MR-pleiotropy residual sum and outliers
- OR:
-
Odds ratio
- SNPs:
-
Single nucleotide polymorphisms
- CXCL9:
-
CXC chemokine ligand 9
- CXCL10:
-
CXC chemokine ligand 10
- YFS:
-
Young Finns study
- b-NGF:
-
Beta nerve growth factor
- CTACK:
-
Cutaneous T-cell attracting chemokine
- FGFBasic:
-
Basic fibroblast growth factor
- G-CSF:
-
Granulocyte colony-stimulating factor
- GRO-a:
-
Growth-regulated oncogene-a
- HGF:
-
Hepatocyte growth factor
- IFNg:
-
Interferon gamma
- IL:
-
Interleukin
- IP-10:
-
Interferon gamma-induced protein 10
- MCP-1:
-
Monocyte chemotactic protein-1
- MCP-3:
-
Monocyte-specific chemokine 3
- M-CSF:
-
Macrophage colony-stimulating factor
- MIF:
-
Macrophage-migration inhibitory factor
- MIG:
-
Monokine induced by interferon gamma
- MIP-1a:
-
Macrophage inflammatory protein-1a
- MIP-1b:
-
Macrophage inflammatory protein-1b
- PDGFbb:
-
Platelet-derived growth factor BB
- RANTES:
-
Regulated on activation, normal T-cell expressed and secreted
- SCF:
-
Stem cell factor
- SCGFb:
-
Stem cell growth factor beta
- SDF-1a:
-
Stromal cell-derived factor-1 alpha
- TNFα:
-
Tumor necrosis factor alpha
- TNFβ:
-
Tumor necrosis factor beta
- TRAIL:
-
TNF-related apoptosis-inducing ligand
- VEGF:
-
Vascular endothelial growth factor
References
Li S, Young KH, Medeiros LJ. Diffuse large B-cell lymphoma. Pathology. 2018;50(1):74–87.
Sehn LH, Salles G. Orcid Id. Diffuse large B-cell lymphoma. N Engl J Med. 2021;384(9):842–58.
Martelli M, Ferreri AJ, Agostinelli C, et al. Diffuse large B-cell lymphoma. Crit Rev Oncol Hematol. 2013;87(2):146–71.
Sant M, Allemani C, Tereanu C, et al. Incidence of hematologic malignancies in Europe by morphologic subtype: results of the HAEMACARE project. Blood. 2010;116(19):3724–34.
Cerhan JR, Kricker A, Paltiel O, et al. Medical history, lifestyle, family history, and occupational risk factors for diffuse large B-cell lymphoma: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monogr. 2014;2014(48):15–25.
Cerhan JR, Berndt SI, Vijai J, et al. Genome-wide association study identifies multiple susceptibility loci for diffuse large B cell lymphoma. Nat Genet. 2014;46(11):1233–8.
Morton LM, Slager SL, Cerhan JR, et al. Etiologic heterogeneity among non-Hodgkin lymphoma subtypes: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monogr. 2014;2014(48):130–44.
Acosta-Rodríguez EV, Merino MC, Montes CL, Motrán CC, Gruppi A. Cytokines and chemokines shaping the B-cell compartment. Cytokine Growth Factor Rev. 2007;18(1–2):73–83.
Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med. 2006;354(6):610–21.
Schaerli P, Moser B. Chemokines: control of primary and memory T-cell traffic. Immunol Res. 2005;31(1):57–74. https://doi.org/10.1385/IR:31:1:572005:57-74.
Bonati L, Tang L. Cytokine engineering for targeted cancer immunotherapy. Curr Opin Chem Biol. 2021;62:43–52.
Yoshimura A, Ito M, Chikuma S, Akanuma T, Nakatsukasa H. Negative regulation of cytokine signaling in immunity. Cold Spring Harb Perspect Biol. 2018;10(7): a028571.
Atallah-Yunes SA, Robertson MJ. Cytokine based immunotherapy for cancer and lymphoma: biology, challenges and future perspectives. Front Immunol. 2022;20(13): 872010.
Shen N, Yu Y, Zhang R, et al. Expression and prognostic value of PIK3CA, VEGF, IL-8, IL-10, and RIP2 in diffuse large B-cell lymphoma. Int J Clin Pract. 2022;7(2022):2637581.
Nacinović-Duletić A, Stifter S, Dvornik S, Skunca Z, Jonjić N. Correlation of serum IL-6, IL-8 and IL-10 levels with clinicopathological features and prognosis in patients with diffuse large B-cell lymphoma. Int J Lab Hematol. 2008;30(3):230–9.
Hashwah H, Bertram K, Stirm K, et al. The IL-6 signaling complex is a critical driver, negative prognostic factor, and therapeutic target in diffuse large B-cell lymphoma. EMBO Mol Med. 2019;11(10): e10576.
Stirm K, Leary P, Bertram K, et al. Tumor cell-derived IL-10 promotes cell-autonomous growth and immune escape in diffuse large B-cell lymphoma. Oncoimmunology. 2021;10(1):2003533.
Nakayama S, Yokote T, Hirata Y, et al. TNF-α expression in tumor cells as a novel prognostic marker for diffuse large B-cell lymphoma, not otherwise specified. Am J Surg Pathol. 2014;38(2):228–34.
Piechna K, Żołyniak A, Jabłońska E, et al. Activity and rational combinations of a novel, engineered chimeric, TRAIL-based ligand in diffuse large B-cell lymphoma. Front Oncol. 2022;31(12):1048741.
Oehadian A, Koide N, Mu MM, et al. Interferon (IFN)-beta induces apoptotic cell death in DHL-4 diffuse large B cell lymphoma cells through tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). Cancer Lett. 2005;225(1):85–92.
Smith GD, Ebrahim S. “Mendelian randomization”: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32(1):1–22.
Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey SG. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27(8):1133–63.
Widding-Havneraas T, Zachrisson HD. A gentle introduction to instrumental variables. J Clin Epidemiol. 2022;149:203–5.
Ahola-Olli AV, Würtz P, Havulinna AS, et al. Genome-wide association study identifies 27 loci influencing concentrations of circulating cytokines and growth factors. Am J Hum Genet. 2017;100(1):40–50.
Kalaoja M, Orcid ID, Corbin LJ, et al. The role of inflammatory cytokines as intermediates in the pathway from increased adiposity to disease. Obesity (Silver Spring). 2021;29(2):428–37.
Burgess S, Orcid ID, Davey Smith G, et al. Guidelines for performing Mendelian randomization investigations: update for summer 2023. Wellcome Open Res. 2020;4:186.
Skrivankova VW, Richmond RC, Woolf BAR, et al. Strengthening the reporting of observational studies in epidemiology using Mendelian randomization: the STROBE-MR statement. JAMA. 2021;326(16):1614–21.
Palmer TM, Lawlor DA, Harbord RM, et al. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat Methods Med Res. 2012;21(3):223–42.
Burgess S, Thompson SG. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40(3):755–64.
Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46(6):1985–98.
Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304–14.
Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–25.
Verbanck M, Chen CY, Orcid ID, et al. Publisher Correction: Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(8):1196.
Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2017;26(5):2333–55.
Burgess S, Orcid ID, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32(5):377–89.
Verbanck M, Chen CY, Orcid ID, et al. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–8.
Hemani G, Orcid ID, Zheng J, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408.
Bao C, Gu J, Huang X, et al. Cytokine profiles in patients with newly diagnosed diffuse large B-cell lymphoma: IL-6 and IL-10 levels are associated with adverse clinical features and poor outcomes. Cytokine. 2023;169: 156289.
Salven P, Orpana A, Teerenhovi L, Joensuu H. Simultaneous elevation in the serum concentrations of the angiogenic growth factors VEGF and bFGF is an independent predictor of poor prognosis in non-Hodgkin lymphoma: a single-institution study of 200 patients. Blood. 2000;96(12):3712–8.
Ruiduo C, Ying D, Qiwei W. CXCL9 promotes the progression of diffuse large B-cell lymphoma through up-regulating β-catenin. Biomed Pharmacother. 2018;107:689–95.
Zhou X, Guo S, Shi Y. Comprehensive analysis of the expression and significance of CXCLs in human diffuse large B-cell lymphoma. Sci Rep. 2022;12(1):2817.
Luster AD, Leder P. IP-10, a -C-X-C- chemokine, elicits a potent thymus-dependent antitumor response in vivo. J Exp Med. 1993;178(3):1057–65.
Ansell SM, Maurer MJ, Ziesmer SC, et al. Elevated pretreatment serum levels of interferon-inducible protein-10 (CXCL10) predict disease relapse and prognosis in diffuse large B-cell lymphoma patients. Am J Hematol. 2012;87(9):865–959.
Moriai S, Takahara M, Ogino T, et al. Production of interferon-{gamma}-inducible protein-10 and its role as an autocrine invasion factor in nasal natural killer/T-cell lymphoma cells. Clin Cancer Res. 2009;15(22):6771–9.
Teichmann M, Meyer B, Beck A, Niedobitek G. Expression of the interferon-inducible chemokine IP-10 (CXCL10), a chemokine with proposed anti-neoplastic functions, in Hodgkin lymphoma and nasopharyngeal carcinoma. J Pathol. 2005;206(1):68–75.
Witzig TE, Maurer MJ, Stenson MJ, et al. Elevated serum monoclonal and polyclonal free light chains and interferon inducible protein-10 predicts inferior prognosis in untreated diffuse large B-cell lymphoma. Am J Hematol. 2014;89(4):417–22.
Noyori O, Komohara Y, Nasser H, et al. Expression of IL-34 correlates with macrophage infiltration and prognosis of diffuse large B-cell lymphoma. Clin Transl Immunology. 2019;8(8): e1074.
Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496(7446):445–55.
Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41(1):49–61.
Nakayama-Ichiyama S, Yokote T, Oka S, et al. Macrophage colony-stimulating factor produced by diffuse large B-cell lymphoma. Br J Haematol. 2010;149(3):310.
Ruddle NH. Lymphotoxin and TNF: how it all began-a tribute to the travelers. Cytokine Growth Factor Rev. 2014;25(2):83–9.
Gommerman JL, Browning JL, Ware CF. The Lymphotoxin Network: orchestrating a type I interferon response to optimize adaptive immunity. Cytokine Growth Factor Rev. 2014;25(2):139–45.
Cao C, Liu S, Lou SF, Liu T. The +252A/G polymorphism in the Lymphotoxin-α gene and the risk of non-Hodgkin lymphoma: a meta-analysis. Eur Rev Med Pharmacol Sci. 2014;18(4):544–52.
Qu Y, Orcid ID, Wang X, et al. The effects of TNF-α/TNFR2 in regulatory T cells on the microenvironment and progression of gastric cancer. Int J Cancer. 2022;150(8):1373–91.
Cruceriu D, Baldasici O, Balacescu O, Orcid ID, Berindan-Neagoe I. The dual role of tumor necrosis factor-alpha (TNF-α) in breast cancer: molecular insights and therapeutic approaches. Cell Oncol (Dordr). 2020;43(1):1–18.
Wang X, Yang L, Huang F, et al. Inflammatory cytokines IL-17 and TNF-α up-regulate PD-L1 expression in human prostate and colon cancer cells. Immunol Lett. 2017;184:7–14.
Ashkenazi A. Directing cancer cells to self-destruct with pro-apoptotic receptor agonists. Nat Rev Drug Discov. 2008;7(12):1001–12.
Amarante-Mendes GP, Griffith TS. Therapeutic applications of TRAIL receptor agonists in cancer and beyond. Pharmacol Ther. 2015;155:117–31.
Wu W, Yang Y, Deng G, et al. Vernodalol enhances TRAIL-induced apoptosis in diffuse large B-cell lymphoma cells. Mol Carcinog. 2017;56(10):2190–9.
Acknowledgements
The authors express their gratitude to the participants and investigators of the FinnGen and YFS, FINRISK studies. We also thank Figdraw (https://www.figdraw.com) for the assistance in model drawing.
Funding
This work was supported by the Youth Project Shaoxing People’s Hospital (Grant Number: 2023YB15), Medical and Health Science and Technology Project of Zhejiang Province (Grant Number: 2023RC107), Department of Health of Zhejiang Province (Grant Number: 2021KY1137), Young Innovative Talents Project of Zhejiang Health Science and Technology Plan (Grant Number: 2022RC078).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Jieni Yu, Chao Xu, and Leihua Fu designed the study, Zhijian Zhang and Lina Ding analyzed and interpreted the data, and drafted the manuscript. Li Hong, Feidan Gao and Jing Jin analyzed and interpreted the data, Jiaping Fu finished the figures, Weiying Feng and Pan Hong revised the manuscript. All author read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
This study has been conducted using published studies and consortia providing publicly available summary statistics. All original studies have been approved by the corresponding ethical review board, and the participants have provided informed consent.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jieni Yu and Leihua Fu are contributed equally to this work and share first authorship.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yu, J., Fu, L., Zhang, Z. et al. Causal relationships between circulating inflammatory cytokines and diffuse large B cell lymphoma: a bidirectional Mendelian randomization study. Clin Exp Med 23, 4585–4595 (2023). https://doi.org/10.1007/s10238-023-01221-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10238-023-01221-y