Abstract
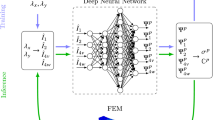
Identifying the constitutive parameters of soft materials often requires heterogeneous mechanical test modes, such as simple shear. In turn, interpreting the resulting complex deformations necessitates the use of inverse strategies that iteratively call forward finite element solutions. In the past, we have found that the cost of repeatedly solving non-trivial boundary value problems can be prohibitively expensive. In this current work, we leverage our prior experimentally derived mechanical test data to explore an alternative approach. Specifically, we investigate whether a machine learning-based approach can accelerate the process of identifying material parameters based on our mechanical test data. Toward this end, we pursue two different strategies. In the first strategy, we replace the forward finite element simulations within an iterative optimization framework with a machine learning-based metamodel. Here, we explore both Gaussian process regression and neural network metamodels. In the second strategy, we forgo the iterative optimization framework and use a stand alone neural network to predict the entire material parameter set directly from experimental results. We first evaluate both approaches with simple shear experiments on blood clot, an isotropic, homogeneous material. Next, we evaluate both approaches against simple shear and uniaxial loading experiments on right ventricular myocardium, an anisotropic, heterogeneous material. We find that replacing the forward finite element simulations with metamodels significantly accelerates the parameter identification process with excellent results in the case of blood clot, and with satisfying results in the case of right ventricular myocardium. On the other hand, we find that replacing the entire optimization framework with a neural network yielded unsatisfying results, especially for right ventricular myocardium. Overall, the importance of our work stems from providing a baseline example showing how machine learning can accelerate the process of material parameter identification for soft materials from complex mechanical data, and from providing an open access experimental and simulation dataset that may serve as a benchmark dataset for others interested in applying machine learning techniques to soft tissue biomechanics.









Similar content being viewed by others
Data Availability
All experimental and synthetic data, as well as all Python code is available for open use under: https://github.com/SoftTissueBiomechanicsLab/ML-soft-material-parameters.
References
Ardizzone L, Kruse J, Wirkert S, Rahner D, Pellegrini EW, Klessen RS, Maier-Hein L, Rother C, Köthe U (2018) Analyzing inverse problems with invertible neural networks. arXiv:1808.04730
Avazmohammadi R, Li DS, Leahy T, Shih E, Soares JS, Gorman JH, Gorman RC, Sacks MS (2018) An integrated inverse model-experimental approach to determine soft tissue three-dimensional constitutive parameters: application to post-infarcted myocardium. Biomech Model Mechanobiol 17(1):31–53
Balaban G, Alnæs MS, Sundnes J, Rognes ME (2016) Adjoint multi-start-based estimation of cardiac hyperelastic material parameters using shear data. Biomech Model Mechanobiol 15(6):1509–1521
Chen CT, Gu GX (2021) Learning hidden elasticity with deep neural networks. Proc Natl Acad Sci 118(31):e2102721118
Costabal FS, Matsuno K, Yao J, Perdikaris P, Kuhl E (2019) Machine learning in drug development: characterizing the effect of 30 drugs on the qt interval using Gaussian process regression, sensitivity analysis, and uncertainty quantification. Comput Methods Appl Mech Eng 348:313–333
Fan R, Sacks MS (2014) Simulation of planar soft tissues using a structural constitutive model: finite element implementation and validation. J Biomech 47(9):2043–2054
Ferruzzi J, Bersi M, Humphrey J (2013) Biomechanical phenotyping of central arteries in health and disease: advantages of and methods for murine models. Ann Biomed Eng 41(7):1311–1330
Frankel AL, Jones RE, Swiler LP (2020) Tensor basis gaussian process models of hyperelastic materials. J Mach Learn Model Comput 1(1):1–17
Frazier PI (2018) A tutorial on Bayesian optimization. arXiv:1807.02811
Gasser TC, Ogden RW, Holzapfel GA (2006) Hyperelastic modelling of arterial layers with distributed collagen fibre orientations. J R Soc Interface 3(6):15–35
Grobbel MR, Shavik SM, Darios E, Watts SW, Lee LC, Roccabianca S (2018) Contribution of left ventricular residual stress by myocytes and collagen: existence of inter-constituent mechanical interaction. Biomech Model Mechanobiol 17(4):985–999
Gu GX, Chen CT, Buehler MJ (2018) De novo composite design based on machine learning algorithm. Extreme Mech Lett 18:19–28
Holzapfel GA, Ogden RW (2009) Constitutive modelling of passive myocardium: a structurally based framework for material characterization. Philos Trans R Soc A Math Phys Eng Sci 367(1902):3445–3475
Holzapfel GA, Gasser TC, Ogden RW (2000) A new constitutive framework for arterial wall mechanics and a comparative study of material models. J Elast Phys Sci Solids 61(1):1–48
Kakaletsis S, Meador WD, Mathur M, Sugerman GP, Jazwiec T, Malinowski M, Lejeune E, Timek TA, Rausch MK (2021) Right ventricular myocardial mechanics: Multi-modal deformation, microstructure, modeling, and comparison to the left ventricle. Acta Biomater 123:154–166
Lejeune E (2021) Geometric stability classification: datasets, metamodels, and adversarial attacks. Comput Aided Des 131:102,948
Lejeune E, Zhao B (2021) Exploring the potential of transfer learning for metamodels of heterogeneous material deformation. J Mech Behav Biomed Mater 117:104,276
Leng Y, Tac V, Calve S, Tepole AB (2021) Predicting the mechanical properties of biopolymer gels using neural networks trained on discrete fiber network data. Comput Methods Appl Mech Eng 387:114,160
Li DS, Avazmohammadi R, Merchant SS, Kawamura T, Hsu EW, Gorman JH, Gorman RC, Sacks MS (2020) Insights into the passive mechanical behavior of left ventricular myocardium using a robust constitutive model based on full 3D kinematics. J Mech Behav Biomed Mater 103:103508. https://doi.org/10.1016/j.jmbbm.2019.103508
Liu M, Liang L, Sun W (2019) Estimation of in vivo constitutive parameters of the aortic wall using a machine learning approach. Comput Methods Appl Mech Eng 347:201–217
Maas SA, Ellis BJ, Ateshian GA, Weiss JA (2012) Febio: finite elements for biomechanics. J Biomecha Eng 134(1):011005
Marchesseau S, Delingette H, Sermesant M, Cabrera-Lozoya R, Tobon-Gomez C, Moireau P, i Ventura RF, Lekadir K, Hernandez A, Garreau M et al (2013) Personalization of a cardiac electromechanical model using reduced order unscented Kalman filtering from regional volumes. Med Image Anal 17(7):816–829
Mihai LA, Budday S, Holzapfel GA, Kuhl E, Goriely A (2017) A family of hyperelastic models for human brain tissue. J Mech Phys Solids 106:60–79
Miller R, Kerfoot E, Mauger C, Ismail TF, Young AA, Nordsletten DA (2021) An implementation of patient-specific biventricular mechanics simulations with a deep learning and computational pipeline. Front Physiol 12:1398
Newville M, Stensitzki T, Allen DB, Ingargiola A (2014) LMFIT: non-linear least-square minimization and curve-fitting for python. https://doi.org/10.5281/zenodo.11813
Ogden RW (1972) Large deformation isotropic elasticity—on the correlation of theory and experiment for incompressible rubberlike solids. Proc R Soc Lond A Math Phys Sci 326(1567):565–584
Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L, Desmaison A, Kopf A, Yang E, DeVito Z, Raison M, Tejani A, Chilamkurthy S, Steiner B, Fang L, Bai J, Chintala S (2019) Pytorch: an imperative style, high-performance deep learning library. In: Wallach H, Larochelle H, Beygelzimer A, dAlché-Buc F, Fox E, Garnett R (eds) Advances in neural information processing systems, vol 32, pp. 8024–8035. Curran Associates, Inc.
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E (2011) Scikit-learn: machine learning in python. J Mach Learn Res 12:2825–2830
Peirlinck M, Sahli Costabal F, Sack K, Choy J, Kassab G, Guccione J, De Beule M, Segers P, Kuhl E (2019) Using machine learning to characterize heart failure across the scales. Biomech Model Mechanobiol 18(6):1987–2001
Pezzuto S, Perdikaris P, Costabal FS (2022) Learning cardiac activation maps from 12-lead ecg with multi-fidelity Bayesian optimization on manifolds. arXiv:2203.06222
Raissi M, Perdikaris P, Karniadakis GE (2019) Physics-informed neural networks: a deep learning framework for solving forward and inverse problems involving nonlinear partial differential equations. J Comput Phys 378:686–707
Rausch M, Karniadakis G, Humphrey J (2017) Modeling soft tissue damage and failure using a combined particle/continuum approach. Biomech Model Mechanobiol 16(1):249–261
Schmid H, Nash MP, Young AA, Röhrle O, Hunter PJ (2007) A computationally efficient optimization kernel for material parameter estimation procedures. J Biomech Eng 129(2):279–283. https://doi.org/10.1115/1.2540860
Schmid H, O’Callaghan P, Nash MP, Lin W, LeGrice IJ, Smaill BH, Young AA, Hunter PJ (2008) Myocardial material parameter estimation: a non-homogeneous finite element study from simple shear tests. Biomech Model Mechanobiol 7(3):161–173. https://doi.org/10.1007/s10237-007-0083-0
Shi L, Yao W, Gan Y, Zhao LY, Eugene McKee W, Vink J, Wapner RJ, Hendon CP, Myers K (2019) Anisotropic material characterization of human cervix tissue based on indentation and inverse finite element analysis. J Biomech Eng 141(9):0910171–0910171
Smith K, Mathur M, Meador W, Phillips-Garcia B, Sugerman G, Menta A, Jazwiec T, Malinowski M, Timek T, Rausch M (2021) Tricuspid chordae tendineae mechanics: insertion site, leaflet, and size-specific analysis and constitutive modelling. Exp Mech 61:19–29
Sugerman GP, Kakaletsis S, Thakkar P, Chokshi A, Parekh SH, Rausch MK (2021) A whole blood thrombus mimic: constitutive behavior under simple shear. J Mech Behav Biomed Mater 115:104,216
Tac V, Costabal FS, Tepole AB (2021a) Automatically polyconvex strain energy functions using neural ordinary differential equations. arXiv:2110.03774
Tac V, Sree VD, Rausch MK, Tepole AB (2021b) Data-driven modeling of the mechanical behavior of anisotropic soft biological tissue. arXiv:2107.05388
Weickenmeier J, Jabareen M, Mazza E (2015) Suction based mechanical characterization of superficial facial soft tissues. J Biomech 48(16):4279–4286
Weickenmeier J, de Rooij R, Budday S, Steinmann P, Ovaert TC, Kuhl E (2016) Brain stiffness increases with myelin content. Acta Biomater 42:265–272
Wu MC, Kamensky D, Wang C, Herrema AJ, Xu F, Pigazzini MS, Verma A, Marsden AL, Bazilevs Y, Hsu MC (2017) Optimizing fluid-structure interaction systems with immersogeometric analysis and surrogate modeling: application to a hydraulic arresting gear. Comput Methods Appl Mech Eng 316:668–693
Zhang X, Garikipati K (2020) Machine learning materials physics: Multi-resolution neural networks learn the free energy and nonlinear elastic response of evolving microstructures. Comput Methods Appl Mech Eng 372:113,362
Acknowledgements
We appreciate support from the National Science Foundation through Grants 2046148 (MKR) and 2127925 (MKR, EL) as well as support from the National Institutes of Health through Grant R21HL161832 and the Office of Naval Research through grant N00014-22-1-2073 (MKR).
Author information
Authors and Affiliations
Corresponding author
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kakaletsis, S., Lejeune, E. & Rausch, M.K. Can machine learning accelerate soft material parameter identification from complex mechanical test data?. Biomech Model Mechanobiol 22, 57–70 (2023). https://doi.org/10.1007/s10237-022-01631-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10237-022-01631-z