Abstract
The development of compact and cost-effective passive treatment systems is of critical importance for acid mine drainage (AMD) remediation in Japan. The purpose of this study was to construct an AMD treatment system comprising a sulfate-reducing bioreactor using rice bran as a carbon source for sulfate-reducing bacteria (SRB) and to demonstrate its stable operation for at least a year in terms of continuous sulfate reduction and metal removal. Our 35 L bioreactor comprised a packed inoculum layer of a mixture of rice husks, limestone, and field soil, which was covered with rice bran. During operation, the AMD input flow rate was adjusted to 11.7 mL/min (hydraulic retention time, HRT; 50 h). Throughout the year, physicochemical analyses of system input and output AMD samples revealed that both pH and oxidation–reduction potential values were consistent with the process of sulfate reduction by SRB, although this reduction was observed to be stronger in summer than in winter. Efficient metal removal was observed, with concentrations at the outlet port of <0.33 mg/L Zn, <0.08 mg/L Cu, and <0.005 mg/L Cd, more than meeting Japan’s national effluent standards. Illumina sequencing of 16S rRNA genes revealed that Desulfatirhabdium butyrativorans-related species, which belong to a lineage within Deltaproteobacteria, were dominant (39–48% of the total SRB population) within the bioreactor.
Zusammenfassung
Die Entwicklung kompakter und kostengünstiger passiver Systeme für die Behandlung von saurem Grubenwasser (sog. Acid Mine Drainage, AMD) ist in Japan von zentraler Bedeutung. Ziel der vorliegenden Studie war die Konstruktion eines AMD-Aufbereitungssystems mit einem sulfatreduzierenden Bioreaktor unter Verwendung von Reiskleie als Kohlenstoffquelle für sulfatreduzierende Bakterien (SRB) und der Nachweis des stabilen Betriebes im Hinblick auf kontinuierliche mikrobielle Sulfatreduktion und Entfernung von Metallen über mindestens ein Jahr. Der 35 L Bioreaktor umfasste eine komprimierte Inokulum-Schicht bestehend aus einer Mischung aus Reishülsen (Spelzen), Kalkstein und Ackerboden überdeckt mit Reiskleie. Im Betrieb wurde der AMD-Zustrom auf 11,7 mL/min eingestellt, was einer Verweilzeit (Hydraulic Retention Time, HRT) von 50 h entspricht. Die physikochemische Analytik von Proben in Systemzufluss und -abfluss über ein Jahr zeigte, dass sowohl pH-Wert als auch Redoxpotential mit dem Prozess der bakteriellen Sulfatreduktion übereinstimmten, wobei eine Verstärkung des Prozesses im Sommer gegenüber dem Winter beobachtet werden konnte. Die Metallfracht wurde erfolgreich reduziert, sodass die im Systemabfluss gemessenen Konzentrationen von <0,33 mg/L für Zink, <0,08 mg/L für Kupfer und <0,005 mg/L für Cadmium die geltenden Einleitkriterien Japans noch deutlich unterschreiten. Mittels Illumina 16S-rRNA-Gen-Sequenzierung konnte gezeigt werden, dass Desulfatirhabdium butyrativorans-verwandte Arten, welche einer Linie innerhalb der Deltaproteobacteria angehören, mit 39-48% der gesamten SRB-Population im Bioreaktor dominant waren
Resumen
El desarrollo de sistemas pasivos de tratamientos compactos y efectivos en costos es de importancia crítica para la remediación de drenaje ácido de minas (AMD) en Japón. El propósito de este estudio fue construir un sistema de tratamiento de AMD que incluía un biorreactor con sulfato-reductoras utilizando salvado de arroz como fuente de carbono para bacteria sulfato-reductoras (SRB) para demostrar su funcionamiento estable por al menos un año en términos de reducción continua de sulfato y remoción de metales. Nuestro biorreactor de 35 L comprendía una capa de inóculo dentro de una mezcla de cáscaras de arroz, caliza y suelo, cubierta con salvado de arroz. Durante la operación, el flujo de AMD se ajustó en 11,7 mL/min (tiempo de retención hidráulico, HRT; 50 h). A lo largo del año, los análisis fisicoquímicos de las muestras de ADM antes y después del sistema de tratamiento, mostraron que los valores de pH y potencial redox fueron consistentes con el proceso de reducción de sulfato por acción de las SRB, aunque esta reducción fue mayor en verano que en invierno. A la salida del sistema, se observó una eficiente remoción de metales con concentraciones menores que 0,33 mg/L Zn, 0,08 mg/L Cu y 0,005 mg/L Cd, que se ajusta a los estándares para efluentes en Japón. La secuenciación del gen 16S rRNA utilizando Illumina reveló que especies relacionadas con Desulfatirhabdium butyrativorans, que pertenece a un linaje dentro de las Deltaproteobacteria, fueron dominantes (39-48% del total de la población de SRB) dentro del biorreactor.
抽象
小型高效酸性矿山废水处理系统研发在日本非常重要。文章旨在建立麦皮为硫酸盐还原菌有机炭源的酸性矿山废水被动处理硫酸盐还原反应器,展示该系统一年多稳定的硫酸盐还原和金属离子去除效果。35升生物反应器内接种层由米糠、石灰石和土壤组成,用麦皮覆盖接种层。在系统运行期间,酸性矿山废水(AMD)输入流量11.7mL/min(水力滞留时间50小时)。流入和流出处理系统的酸性矿山废水年物理化学分析说明:即使夏天还原过程较冬天强烈,pH值和氧化-还原电位值仍与硫酸盐还原菌的还原过程总体一致。系统表现出良好的金属去除效果,流出液浓度(锌< 0.33 mg/L、铜< 0.08 mg/L和镉< 0.005 mg/L)远远超过日本排放废水要求。16S rRNA基因的Illumina序列说明,Deltaproteobacteria系Desulfatirhabdium butyrativorans种占总生物反应器硫酸盐还原菌落39-48%
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acid mine drainage (AMD) from metal and coal mines is a significant environmental concern if it is discharged untreated into rivers (Colmer and Hinkle 1947; Price and Errington 1998). Therefore, many treatment methods have been used to neutralize and remove harmful metals from AMD (Rakotonimaro et al. 2016).
In Japan, there are many abandoned mine sites where metal-containing AMD occurs. Municipalities, governments, and companies that hold mining rights are responsible for treating these effluents. The business of AMD treatment is non-profit; therefore, cost savings are required where possible. Most AMD treatment methods at mine sites in Japan have used active treatment processes with chemical reagents, e.g. neutralization and sedimentation, because such approaches can shorten the processing time and are easily controllable. However, active treatment processes bear significant costs to cover the continuous maintenance of facilities, the addition of alkalinity, and the input of manpower. Indeed, in Japan, the treatment of AMD effluents costs ≈$37.8 million per year (Mine Safety and Explosives Control Division, METI, Japan 2013).
By contrast, passive and semi-passive treatment processes are less expensive and have less maintenance demands. One sustainable approach for removing metals from AMD is the use of biochemical reactors (bioreactors). Bioreactors with sulfate-reducing bacteria (SRB) offer particular advantages, such as the production of a compact sludge, low operation costs, and minimal maintenance costs (Gusek et al. 1998). In certain countries other than Japan, full-scale passive treatment facilities using SRB have been introduced. These typically have very large surface areas, allowing a long hydraulic retention time (HRT; Nairn et al. 2010; The ITRC Biochemical Reactors for Mining-Influenced Waste Team 2013). However, it is difficult to build large facilities for passive AMD treatment in Japan because many mine sites are located in steep mountainous areas. Therefore, to be used in Japan, compaction and optimization of the processes with reduced HRT is necessary.
Japan Oil, Gas and Metals National Corporation (JOGMEC) has been developing a compact passive treatment process with a vertical flow bioreactor that contains SRB (Hamai et al. 2016). Since the concentration of dissolved organic carbon in AMD is very low (chemical oxygen demand is usually less than 10 mg/L of O2; Kolmert and Johnson 2001), a carbon source must be added to the bioreactor to promote SRB activity. To reduce operation costs, organic wastes such as animal manure, saw dust, wood chips, rice straw, and corn husks have been used as carbon sources for SRB (Chang et al. 2000; Gibert et al. 2004; Hiibel et al. 2011; McCullough and Lund 2011; Neculita et al. 2007, 2011; Zagury et al. 2006; Zhang and Wang 2014). Among such organic wastes, rice bran, which contains starches, lipids, proteins, and other polysaccharides, is thought to be easily degraded to simple carbon substrates with low molecular weights that are suitable for SRB utilization. Therefore, JOGMEC is assessing the novel application of rice bran as a source of microbial carbon and nitrogen in bioreactors.
In the present study, a passive sulfate-reducing bioreactor using rice bran and rice husks (JOGMEC process) was operated for a year. Water quality within the bioreactor (temperature, pH, oxidation–reduction potential (ORP), and sulfate, sulfide, and metal concentrations) were continually monitored. Additionally, the SRB community present at around 5 °C, the lowest AMD temperature observed in this study, was analyzed by Illumina sequencing.
Materials and Methods
Acid Mine Drainage (AMD) and Iron Removal
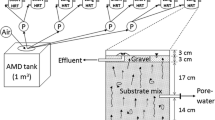
The raw AMD (pH 3.5) used in this study contained approximately 40 mg/L Fe (as ferrous iron), 15 mg/L Zn, 5 mg/L Cu, and 0.06 mg/L Cd, as well as 300 mg/L of sulfate. The AMD was initially passed through an iron-oxidation removal reactor, which reduced the iron concentration to <5 mg/L (pH 3.0). The reactor discharge was then fed into a sulfate-reducing bioreactor using an electric pump (Fig. 1).
Treatment flow of the JOGMEC process. The raw AMD ferrous ion concentration of 40 mg/L was reduced to <5 mg/L by a pre-treatment process in an iron-oxidation removal reactor; the pH value was also decreased to around 3.0 from 3.5. Pre-treated AMD was then fed into the sulfate-reducing bioreactor using an electric pump
Passive Sulfate-Reducing Bioreactor
The reactor was constructed using a polyvinyl chloride column (1100 mm height × 250 mm diameter; Fig. 1) in a prefabricated laboratory at an abandoned mine site. The raw AMD flowed into the reactor through an inlet port at the top of the column, and the treated AMD flowed through an outlet at the base. The reactor had four ports that allowed for sampling of the AMD at different column heights (every 250 mm from the bottom; Fig. 1). The column was first packed with an inoculum mixture of 4.5 kg of rice husks, 18 kg of limestone (3–20 mm in particle diameter), and 17.5 g of field soil in a layer of approximately 1000 mm. Then, 1.5 kg of rice bran was placed on top of this layer before the column was sealed and subsequently saturated with approximately 35 L of AMD, which flowed through the reactor at a prescribed HRT. This passive AMD treatment system, using a sulfate-reducing bioreactor consisting of rice bran and rice husks, was termed the “JOGMEC process.” The input flow rate was adjusted to 11.7 mL/min (HRT: 50 h) using an EHN-B11VHMR electric pump (IWAKI).
Sampling and Chemical Analyses of Treated AMD from the Bioreactor
The experiment was initiated in September 2013 and sampling continued until the end of January 2015. Water from the reactor was sampled from one to several times per week at the inlet port, outlet port, and four sampling ports (ports 1–4 in Fig. 2). The sulfate ion concentration in the sample water was analyzed using a Dionex ICS-2100 ion chromatography system with a Dionex IonPac AS22 column (Thermo Scientific). The pH, ORP, and temperature were measured using a LAQUA act D-73 pH/Ion meter (HORIBA Scientific). For metal ion concentration analysis, 25 mL of sample was desiccated on a heated sand bath after the addition of 1 mL of nitric acid and 50 µL of 60 wt% of perchloric acid to break down any organic matter. The dried sample was then dissolved in 185 µL of nitric acid and diluted to 25 mL. The Zn, Cu, and Cd concentrations were determined using an SPS3100 plasma spectrometer (SII Nano Technology) according to the “General Rules for Atomic Emission Spectrometry” (JIS K 0116:2014). Monitoring of the Zn and Cu concentrations in the bioreactor water samples was initiated in September 2013 (at start-up), whereas for Cd it was started in March 2014.
Conceptual diagram (a) and appearance (b) of a vertical flow sulfate-reducing bioreactor that uses rice bran and rice husks to treat acid mine drainage [Japan Oil, Gas and Metals National Corporation (JOGMEC) process]. The bioreactor was set up in a prefabricated laboratory and covered in insulation material. The laboratory temperature was not artificially controlled
SRB Community Analysis by Illumina Sequencing
Samples for microbial community analyses were obtained from the bottom (sampling Port 4) of the sulfate-reducing bioreactor in January 2015 (day 492). DNA extraction, 16S rRNA gene amplification, Illumina sequencing, and sequence data analyses were carried out by Nippon Steel & Sumikin Eco-Tech. Briefly, DNA was extracted from the samples using the ISOIL for beads beating kit (Nippon Gene), quantified using the PicoGreen dsDNA assay kit (Invitrogen), and used as a template for polymerase chain reaction (PCR) amplification. The V4–V5 region of 16S rRNA genes was amplified using the primers U515F (sequence: GTGYCAGCMGCCGCGGTA) and 926R (sequence: CCGYCAATTCMTTTRAGTT). Appropriate amounts of the 16S rRNA gene segments were subjected to paired-end sequencing with a MiSeq sequencer (Illumina). Removal of low-quality and chimeric sequences, as well as operational taxonomic unit (OTU) grouping (97% sequence identity cut-off), were performed using the QIIME software package (Caporaso et al. 2010). Representative sequences for each OTU were assigned using the Greengene (DeSantis et al. 2006) and SILVA software packages (http://www.arb-silva.de/projects/living-tree/). A phylogenetic tree diagram of the SRB identified in this study was drawn based on the neighbor-joining method (Saitou and Nei 1987) using MEGA7 software (ver. 7.0.18; Kumar et al. 2016). Evolutionary distances were computed using the maximum composite likelihood method (Tamura et al. 2004).
Results and Discussion
Seasonal Changes in Temperature, pH, and ORP
The location of the experimental site was in northern Japan. The average temperature in the prefabricated laboratory during the summer season (May–October) was 18.8 °C, whereas in the winter (November–April), it was 5.6 °C. The temperature of the raw AMD was relatively stable between 10 and 16 °C (12.9 °C average) throughout the year (Fig. 3a). However, as the laboratory room temperature was not controlled, the temperature of the AMD within the column at times fell below 10 °C in winter (for example, in mid-November 2013). The lowest temperature recorded was approximately 5 °C from January to February 2014 (Fig. 3a).
Seasonal changes in temperature (a), pH (b), and oxidation–reduction potential (ORP) (c) in the sulfate-reducing bioreactor from September 2013 to January 2015. Symbols: open circle Port 1, open triangle Port 2, open diamond Port 3, open square Port 4, cross outlet, closed circle acid mine drainage (AMD)
Regarding the pH of the AMD in the bioreactor (originally pH 3.5), an increasing tendency was observed over time (Fig. 3b). Initial neutralization of the AMD occurred in the bioreactor due to the limestone. As a result, the pH increased to the optimum range (5.0–9.0) for SRB activity (Postgate 1984). The pH of the sampled water subsequently remained unchanged at around 7.0 throughout the remainder of the year (except at Port 1, which was located nearest to the bioreactor inlet [Fig. 2] and so was subject to only a short interaction period with the limestone). Until mid-November 2013, the pH of water from Port 1 continued to decrease until it was ≈5.0; this pH was maintained from January to May 2014. Subsequently, the pH of the sampled water increased to around pH 6.5 at Port 1, before decreasing again in October 2014 (Fig. 3b). Bacterial dissimilatory reduction of the sulfate ion to the sulfide ion generates alkalinity (as the hydrogen carbonate ion), as indicated in Formula [1]. In other words, it is believed that the SRB provided pH buffering. As SRB in the bioreactor actively catalyzed sulfate ion reduction, especially during the summer, system pH increased (Fig. 3b).
ORP values favorable for SRB (less than −100 mV; Postgate 1984) were achieved in the bioreactor throughout the year (Fig. 3c). Even more suitable ORP values of −300 mV (Prasad et al. 1999) were achieved during the summer, and in most cases, bioreactor ORP values did not rise above −200 mV, except at Port 1, where it ranged from −240 to −39 mV from mid-November 2013 to April 2014, and from −274 to +77 mV from October 2014 to January 2015. The decrease in ORP during the summer season was probably due to oxygen consumption associated with the microbial degradation of the rice bran. Overall, both pH and ORP were favorable for bioreactor SRB.
Variations in Sulfate Reduction and Sulfide Generation Over Time
Initial sulfate ion concentrations at the inlet port ranged from 245 to 380 mg/L (average 300 mg/L). As shown in Fig. 4a, sulfate reduction was observed even in the uppermost portion of the reactor, i.e. the sulfate concentrations of water sampled from Port 1 were about 25–50% of those at the inlet port. The sulfate ion concentrations at Port 1 were initially around 230 mg/L (September to mid-November 2013), but rose above 250 mg/L between mid-November 2013 and April 2014. After May 2014, the sulfate ion concentrations decreased to around 150 mg/L, but they increased again to above 250 mg/L at the beginning of October 2014 (Fig. 4a). Based on this trend, it appears that the SRB reduced the AMD sulfate ions to sulfide ions using low molecular carbon sources, such as lactate, ethanol, and other organic materials, as electron donors. According to the reaction [1] (Neculita et al. 2007), these low molecular carbon sources (shown as “CH2O”) would be generated from the digested rice bran in the reactor during the JOGMEC process.
The variations in sulfate ion concentrations were consistent with the observed trend in ORP values at Port 1, where ORP values rose above −150 mV [ranging from −240 to −39 mV from mid-November 2013 to April 2014, and from −274 to +77 mV from October 2014 to January 2015 (Fig. 3c)].
The effluent sulfate concentrations at the outlet port ranged from 80 to 276 mg/L (Fig. 4a). Sulfate was stable at ≈200 mg/L until April 2014. After that, a drastic decrease in sulfate concentration (to below 100 mg/L) was observed during the summer, before the concentration gradually increased (to around 250 mg/L) until mid-October 2014. This is consistent with the fact that the highest ORP values at the outlet port were measured from October 2014 to January 2015 (−291 to −84 mV; Fig. 3c). The highest sulfate ion reduction rate (77.8%) was observed in September 2014, whereas the lowest (0%) was observed in January 2015.
Figure 4b shows the sulfide ion concentrations of sampled waters at Ports 1–4 of the bioreactor. Sulfide ion concentrations increased at the lower port locations. From May 2014 to September 2014, when sulfate-reducing activity was relatively high, the sulfide ion concentrations of the water sampled at Port 4 ranged from 19 to 57 mg/L (Fig. 4b). By contrast, when the sulfate-reducing activity was relatively low, the sulfide concentrations were 0–14 mg/L. In Fig. 4, the relationship between changes in (a) and (b) is consistent with sulfate reduction causing sulfide ion generation. According to the reaction given in Formula [2] (Neculita et al. 2007), the sulfide ion in solution is available to precipitate metals (shown as “M” in Formula [2]).
As such, metal sulfides are immobilized within the reactor; thus, metal removal from the AMD is expected.
Metal Removal
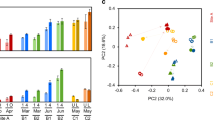
Before treatment in the bioreactor, the iron concentration in the AMD was reduced to <5 mg/L by an iron-oxidation reactor (initial average: 40 mg/L). This pretreated water was fed into the sulfate-reducing bioreactor. Metal (Zn, Cu, and Cd) removal efficiencies by the bioreactor were generally high (Fig. 5).
Variations in metal removal during AMD-impacted water treatment using the sulfate-reducing bioreactor from September 2013 to January, 2015. Shown are the concentrations of Zn (a), Cu (b), and Cd (c). Concentration values show the total concentration of the soluble ion in water and the precipitated ion in suspended precipitates. Symbols: open circle Port 1, open triangle Port 2, open diamond Port 3, open square Port 4, cross outlet, closed circle AMD
From September 2013 to September 2014, most of the Zn was removed in the upper portion of the bioreactor. Removal rates of Zn at Ports 1 and 2 were 83.6–100% (except on sampling day 141; Fig. 5a). However, Zn concentrations became unstable and increased after November 2014. A potential reason for this phenomenon is the decrease in sulfate-reducing activity at the lower temperatures observed during winter. However, considering that the Zn removal rate was relatively stable during the first winter (2013–2014), the depletion of rice bran during continuous operation throughout the year [leading to a lack of organic substrates for SRB in the second winter (2014–2015)] might have caused the decreased sulfate-reducing activity. This hypothesis is supported by the higher ORP values observed in the second winter than in the first winter (Fig. 3c). By contrast, at Port 4, Zn concentrations of <0.33 mg/L were observed throughout the year. These values were below Japan’s national effluent standard for Zn (2 mg/L).
The bioreactor was also effective at Cu removal. The Cu concentrations at the inlet port ranged from 3.2 to 10.0 mg/L (Fig. 5b), whereas effluent concentrations were <0.08 mg/L. Therefore, Cu removal rates were 99.1–100%. In addition, although Cd measurements were only started in March 2014, the trend in Cd removal by the bioreactor was observed to be similar to that of Cu (Fig. 5c). The results showed 93.3–100% removal rates and the effluent concentrations were <0.005 mg/L. The trends in metal removal in the bioreactor corresponded with sulfate ion removal and sulfide ion generation, strongly suggesting metal sulfide precipitation.
Analysis of SRB Populations within the Bioreactor in January
To provide low molecular organic substrates, e.g. lactic acid, acetic acid, formic acid, and ethanol, to SRB, efficient digestion and fermentation of polysaccharides in the rice bran by environmental microorganisms is necessary. SRB and fermentative bacteria are believed to construct ecosystems in the bioreactor, but little is known about the SRB communities that treat AMD. The recent development of high-throughput DNA sequencing technologies, especially the Illumina platform for 16S rRNA amplicon sequencing, enables analysis of the complicated microbial community structures in the environment (Hori et al. 2015; Sato et al. 2016); thus, SRB communities in the bioreactor could be monitored at a fine scale.
The seasonal changes in all measured physicochemical parameters suggested that microbial activity in the bioreactor was generally higher in summer than in winter. Therefore, in the summer, the higher efficiency of metal removal and sulfate ion reduction was likely achieved due to both higher microbial degradation of the rice bran and greater proliferation of the SRB population, even in the upper portion of the reactor (around Port 1).
On the other hand, with the onset of winter, a gradual decrease in sulfate-reducing activity within the bioreactor, followed by an increase in ORP values, was observed (Fig. 4a). The lowest sulfate ion reduction rates were observed in January, when the laboratory temperature was low (Fig. 3a). To better understand SRB populations during the cold season, the most dominant SRB genus within the bioreactor (in samples from Ports 1 and 4) was investigated in January 2015 (506 days) by Illumina sequencing of 16S rRNA genes. The total numbers of 16S rRNA gene sequences obtained from Ports 1 and 4 were 128,527 and 123,927, respectively; SRB were found to dominate, comprising 40.5 and 10.4% of all sequences, respectively. This result indicated that the SRB population was greater at Port 1 than it was at Port 4. Of the SRB population at both sampling ports, Desulfatirhabdium butyrativorans-related species belonging to a novel lineage within Deltaproteobacteria (Balk et al. 2008) dominated, accounting for 47.7 and 39.4% of the total SRB communities in Ports 1 and 4, respectively (19.3 and 4.1% of the total number of 16S rRNA gene sequences, respectively). The phylogenetic affiliations of Deltaproteobacteria-related SRB found at Ports 1 and 4 are presented in Supplemental Fig. 1. The presence of nutritionally versatile SRB affiliated with Deltaproteobacteria, as well as another type of SRB affiliated with Firmicutes (data not shown), was observed within the bioreactor, suggesting that the digestion and fermentation of rice bran resulted in a diverse range of organic products.
Conclusion
A laboratory-scale sulfate-reducing bioreactor containing rice bran and husk (both agricultural wastes) was successfully operated to treat AMD for over a year. Efficient metal removal was observed, with concentrations at the outlet port of <0.33 mg/L Zn, <0.08 mg/L Cu, and <0.005 mg/L Cd during operation. These values more than meet Japan’s national effluent standards. Metabolically diverse SRB populations within the bioreactor in January were identified by Illumina sequencing. In particular, it was revealed that Desulfatirhabdium butyrativorans-related species belonging to a lineage within Deltaproteobacteria dominated (39–48% of the total SRB population) within the bioreactor. Considering that a sulfidogenic condition was maintained throughout the operation of the bioreactor, SRB could likely use simple carbon substrates derived from the microbial digestion and fermentation of the rice bran. Therefore, the JOGMEC treatment process is potentially an effective metal removal process for AMD.
References
Balk M, Altinbas M, Rijpstra WI, Sinninghe Damsté JS, Stams AJ (2008) Desulfatirhabdium butyrativorans gen. nov., sp. nov., a butyrate-oxidizing, sulfate-reducing bacterium isolated from an anaerobic bioreactor. Int J Syst Evol Microbiol 58:110–115
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336
Chang IS, Shin PK, Kim BH (2000) Biological treatment of acid mine drainage under sulfate-reducing conditions with solid waste materials as substrate. Water Res 34(4):1269–1277
Colmer AR, Hinkle ME (1947) The role of microorganisms in acid mine drainage: a preliminary report. Science 106(2751):253–256
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72(7):5069–5072
Gibert O, de Pablo J, Cortina JL, Ayora C (2004) Chemical characterization of natural organic substrates for biological mitigation of acid mine drainage. Water Res 38(19):4186–4196
Gusek J, Wildeman T, Miller A, Fricke J (1998) The challenges of designing, permitting and building a 1,200 gpm passive bioreactor for metal mine drainage West Fork mine, Missouri. In: Proceedings of 15th Annual Meeting of the American Soc for Surface Mining and Reclamation (ASSMR), St. Louis, MO, pp. 203–212
Hamai T, Kodera T, Kobayashi M, Masuda N, Sakata T (2016) The sequential tests about passive treatment for acid mine drainage using sulfate reducing bacteria (In Japanese). J MMIJ 132(11):175–181
Hiibel SR, Pereyra LP, Riquelme Breazeal MV, Reisman DJ, Reardon KF, Pruden A (2011) Effect of organic substrate on the microbial community structure in pilot-scale sulfate-reducing biochemical reactors treating mine drainage. Environ Eng Sci 28(8):563–571
Hori T, Aoyagi T, Itoh H, Narihiro T, Oikawa A, Suzuki K, Ogata A, Friedrich MW, Conrad R, Kamagata Y (2015) Isolation of microorganisms involved in reduction of crystalline iron(III) oxides in natural environments. Front Microbiol 6:386. doi:10.3389/fmicb.2015.00386/full
Kolmert Å, Johnson DB (2001) Remediation of acidic waste waters using immobilised, acidophilic sulfate-reducing bacteria. J Chem Technol Biotechnol 76(8):836–843
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis, version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
McCullough CD, Lund MA (2011) Bioremediation of acidic and metalliferous drainage (ADM) through organic carbon amendment by municipal sewage and green waste. J Environ Manage 92:2419–2426
Mine Safety and Explosives Control Division, Commerce, Distribution and Industrial Safety Policy Group (2013) Act on special measures for pollution caused by the metal mining industry, etc. Policy Index of Ministry of Economy, Trade and Industry, Japan
Nairn RW, LaBar JA, Strevett KA, Strosnider WH, Morris D, Neely CA, Garrido A, Santamaria B, Oxenford L, Kauk K, Carter S, Furneaux B (2010) A large, multi-cell, ecologically engineered passive treatment system for ferruginous lead-zinc mine waters. In: Proceeding of international mine water association annual meeting, pp 21–24
Neculita CM, Zagury GJ, Bussiere B (2007) Passive treatment of acid mine drainage in bioreactors using sulfate-reducing bacteria: critical review and research needs. J Environ Qual 36:1–16
Neculita CM, Yim GJ, Lee G, Ji SW, Jung JW, Park HS (2011) Comparative effectiveness of mixed organic substrates to mushroom compost for treatment of mine drainage in passive bioreactors. Chemosphere 83:76–82
Postgate JR (1984) The sulphate reducing bacteria, second edit. Cambridge University Press, Cambridge
Prasad D, Wai M, Berube P, Henry JG (1999) Evaluating substrates in the treatment of acid mine drainage. Environ Technol 20:449–459
Price WA, Errington JC (1998) Guidelines for metal leaching and acid rock drainage at minesites in British Columbia. Ministry of Energy and Mines, British Columbia
Rakotonimaro TV, Neculita CM, Bussière B, Benzaazoua M, Zagury GJ (2016) Recovery and reuse of sludge from active and passive treatment of mine drainage-impacted waters: a review. Environ Sci Pollut Res 24(1):73–91
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Sato Y, Hori T, Navarro RR, Habe H, Ogata A (2016) Functional maintenance and structural flexibility of microbial communities perturbed by simulated intense rainfall in a pilot-scale membrane bioreactor. Appl Microbiol Biotechnol 100(14):6447–6456
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci 101(30):11030–11035
The Interstate Technology & Regulatory Council (ITRC) Biochemical Reactors for Mining-Influenced Waste Team (2013) Biochemical reactors for mining-influenced water. http://itrcweb.org/bcr-1/
Zagury GJ, Kulnieks VJ, Neculia CM (2006) Characterization and reactivity assessment of organic substrates for sulfate-reducing bacteria in acid mine drainage treatment. Chemosphere 64:944–954
Zhang M, Wang H (2014) Organic wastes as carbon sources to promote sulfate reducing bacterial activity for biological remediation of acid mine drainage. Miner Eng 69:81–90
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Sato, Y., Hamai, T., Hori, T. et al. Year-Round Performance of a Passive Sulfate-Reducing Bioreactor that Uses Rice Bran as an Organic Carbon Source to Treat Acid Mine Drainage. Mine Water Environ 37, 586–594 (2018). https://doi.org/10.1007/s10230-017-0489-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10230-017-0489-6