Abstract
The striped eel catfish, Plotosus lineatus was first described by Thunberg in 1787 from the Indo-Pacific region in the East Indian Ocean. Since then, the species has been recorded in various marine and brackish habitats in Japan, southern Korea, the Ogasawara Islands, Australia, Lord Howe Island, Palau and Yap in Micronesia, East Africa to Samoa, Madagascar, Red Sea and the Persian Gulf. Occurrences of this species have also been registered in the Mediterranean Sea, a non-native location, indicating a possible biological invasion event. Despite its long history, the taxonomic status of the P. lineatus species complex remains puzzling and uncertain. Here, we analysed all the available mitochondrial cytochrome c oxidase subunit 1 sequences (NCBI and BOLD) from specimens covering the current known distribution range, in an attempt to clarify the evolutionary relationships of different lineages within this species. We deduce nine monophyletic Lineages - I–IX, of P. lineatus with Kimura-2P distances ranging from 2% to 16%, with a mean intraspecific distance of 6%. Strikingly, Lineage V is composed uniquely of Mediterranean-captured individuals, with an unknown evolutionary origin. These findings strongly suggest the need for a careful species taxonomic reassessment. Some Lineages are composed of individuals from specific geographic locations (e.g., Australia and Indonesia), while others include specimens from broader geographic areas (e.g., almost all Indo-Pacific coastline). Additionally, several deposited sequences are most likely the result of morphology-based misidentifications. Due to the biological invasive potential, as well as the use of the species as a valuable physiology model, the P. lineatus species complex requires further attention. Overall our study offers a clear framework for future comparative studies of striped eel catfish individuals captured from different ecosystems.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The eel catfish genus Plotosus Lacepède 1803 is widely distributed in the tropical Indo-West Pacific, with species occurring in estuarine, coastal shallow waters and offshore deeper waters habitats (e.g., Gomon and Taylor 1982; Burgess 1989; Ferraris 1999; Ng and Sparks 2002). The number of valid species in this genus is highly contentious, but it includes at least nine recognized species: the dog-tooth eel catfish Plotosus canius Hamilton 1822, the short-tail eel catfish Plotosus abbreviatus (Boulenger 1895), Plotosus fisadoha Ng and Sparks 2002, the Japanese eel catfish Plotosus japonicus Yoshino and Kishimoto 2008, the spiny eel catfish Plotosus limbatus Valenciennes in Cuvier and Valenciennes 1840, the striped eel catfish Plotosus lineatus (Thurnberg 1787), Plotosus nhatrangensis Prokofiev 2008, the pond spiny eel catfish Plotosus nkunga Gomon and Taylor 1982 and Plotosus papuensis Weber 1910. The latter is the only species to be found in freshwater habitats (Fricke and Eschmeyer 2023; Froese and Pauly 2023).
The type species of the genus, Plotosus lineatus, commonly known as striped eel catfish or lined catfish, is eel-shaped, with a brown body and two white longitudinal stripes and was described from the Indo-Pacific region, in the East Indian Ocean, more than 200 years ago (Plotosus lineatus Thunberg 1787). It has the widest distribution range of all species of Plotosus. Geographically, it has been collected in Japan, southern Korea, the Ogasawara Islands, Australia, Lord Howe Island, Palau and Yap in Micronesia, East Africa to Samoa, Madagascar, Red Sea and the Persian Gulf (Taylor and Gomon 1986; Myers 1999; Wakwabi and Mees 1999; Ali et al. 2007; Ketabi and Jamily 2016; Froese and Pauly 2023).
Since the opening of the Suez Canal in 1869, about 100 fish species have migrated from the Red Sea to the Mediterranean Sea (Bentur et al. 2018). Plotosus lineatus was recorded for the first time in the Mediterranean Sea in 2002 (in Israel), as the second introduced catfish species (Golani 2002). Since then, several other occurrences in the Mediterranean followed, with large schools being observed and bycatch being present in fishing nets in Egypt (Temraz and Souissi 2013), Lebanon (Bitar 2013), Syria (Ali et al. 2015), Tunisia (Ounifi-Ben Amor et al. 2016), and Turkey (Bayhan and Ergüden 2022). Despite its relatively slow rate of spread in the area, it has been alleged to displace native fishes through competition (Edelist et al. 2012) and is included in the list of the 100 worst invasive species in the Mediterranean (Streftaris and Zenetos 2006). Plotosus lineatus was assessed as a high-risk marine invasive alien species for the EU and included in the list of invasive alien species of Union concern (the Union list) due to its potential to threaten native ecosystems and human well-being throughout the Mediterranean and within EU marine waters (Galanidi et al. 2019; http://data.europa.eu/eli/reg_impl/2016/1141/oj).
To date, the taxonomic status of P. lineatus remains unclear. This taxonomic clarification obstacle is worrisome since the species is important for both the aquarium trade and human consumption (Situ and Sadovy 2004; Asriyana et al. 2021, 2022). Moreover, it is also an insightful model organism to study osmoregulation, given its uniqueness among teleosts by possessing a dendritic organ (e.g., Lanzing 1967; Malakpour Kolbadinezhad et al. 2018a, b) and ammonia excretion (Malakpour Kolbadinezhad et al. 2020). Additionally, P. lineatus has venomous spines that cause painful injuries, sometimes accompanied by hypertension and tachycardia and a risk of secondary infection (Dogdu et al. 2016; Bentur et al. 2018), which has serious socio-economic effects in the area and, for example, forces shrimp fishermen to change the time and place of fishing (Edelist et al. 2012; Galanidi et al. 2018). Moreover, it has been widely used as a model organism to study the potential use of its venom in biomedical applications (Shiomi et al. 1988; Fahim et al. 1996). Recently, a reference nuclear genome assembly has been released (Shao et al. 2023). Given the complexity of the taxonomic status of Plotosus, a comprehensive phylogenetic study of the various lineages and diversity will have wide impacts on future research avenues of this genus.
Few attempts have been made to clarify the genetic diversity within the species, but some studies have suggested the existence of several evolutionary lineages within P. lineatus (Bariche et al. 2015; Kundu et al. 2019; Goren et al. 2020). Nevertheless, these studies were limited either in geographic or phylogenetic scope. No study has ever included P. lineatus individuals from across its full (native and introduced) distributional range, and using all the available mtDNA Cytochrome c oxidase subunit 1 (COI) sequences, which is a standardized molecular identification system used in fishes (Hebert et al. 2003). Although is a single mtDNA marker, the effectiveness and benefits of barcoding fishes have been demonstrated in flagging previously overlooked species and enabling identifications where traditional methods cannot be applied, or are insufficient (Ward et al. 2009).
In this study, all available Plotosus sp. mitochondrial COI sequences (NCBI and BOLD; n = 204) were analysed, including all P. lineatus from specimens covering the current known distribution range, in an attempt to clarify the diversity of lineages in this species. We discuss several aspects that have added to P. lineatus taxonomic uncertainty. Finally, to prevent the recurrence of the major problems observed, we created a comprehensive list of all information accessible for each sequence, including morphology-based misidentifications and the place where the samples were collected, both of which are important details that are not always (easy) directly related to the sequence resources.
Materials and methods
Phylogenetic analyses. For the phylogenetic analysis, all sequences labelled as Plotosus were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/genbank/) and the Barcode of Life Data System (BOLD) (accessed in January of 2023) [Electronic Supplementary Material (ESM) Table S1]. Redundant entries and sequences with less than 500 bp were manually removed (i.e., sequences KP296155.1 and MH331830.1). Several Gymnotiformes and one representative of each Siluriformes family (according to Betancur-R et al. 2017) were also retrieved from NCBI and used as outgroup (see ESM File S1 for the Accession numbers).
Alignments were produced using MAFFT v7.453 (Katoh and Standley 2013) (parameters: default) and trimmed after with trimAL v.1.2rev59 (Capella-Gutiérrez et al. 2009) (parameters: -gt 0.5). The final alignment consisted of 237 sequences and a total length of 645 bp. The resulting alignment was translated to infer putative indels and unexpected stop codons. The best-fit model of nucleotide substitution evolution and Maximum Likelihood (ML) phylogenetic inference were produced in IQ-TREE v.1.6.12 (Nguyen et al. 2015; Kalyaanamoorthy et al. 2017).
Genetic diversity and haplotype network. For each of the major clades identified in the phylogeny, genetic distances (Kimura 2-parameter distance model, K2P) were assessed using MEGA11 (Kumar et al. 2018). For haplotype network construction, all sequences placed in the P. lineatus cluster (depicted in the phylogeny, see Results) were retrieved from the alignment and trimmed with trimAL v.1.2rev59 (Capella-Gutiérrez et al. 2009) (parameters: -gt 0.95) to a length of 564 bp. The haplotype network was inferred using TCS 2.1 (Clement et al. 2000) implemented through PopART (Leigh and Bryant 2015), considering sequences with the same length (407 bp).
Plotosus lineatus lineage distribution. Based on published and unpublished data, sampling locations for all the sequences used to construct the haplotype network were gathered and assembled (ESM Table S2). The acquired coordinates were loaded into QGis 3.22, whereas for the remaining sequences/haplotypes for which only approximations of places were obtained (e.g., Country and/or Province name), an approximation of location was represented using the same software. The acquired P. lineatus lineages distributions were then depicted using the same haplotype network's colour scheme (and phylogeny).
Results
Plotosus spp. phylogenetic relationships. In the COI alignment used for the phylogenetic inference, no stop codon, insertions or deletions were found in any of the sequences. The ML phylogeny retrieves one highly supported clade, Plotosidae, which includes all Plotosus spp. sequences except for two that cluster with the Nematogenyidae representative, very likely resulting from morphology-based misidentifications (Plotosus sp. UKFBJ138-08 and KF268174.1 in Fig. 1; ESM Table S1).
DOWN: maximum likelihood (ML) phylogenetic tree inferred from the cytochrome c oxidase subunit I (COI) gene fragment for one representative of each Siluriformes family, all Plotosidae sequences available (collapsed for easier visualization) and several Gymnotiformes; Asterisk indicate unknown clustering of two sequences deposited as Plotosus lineatus. For details see ESM Table S1 and ESM File 1. TOP: Uncolapsed Plotosidae Clade. Only bootstrap values > 60% are depicted. TOP-RIGHT: photographs of all available individuals retrieved from the Barcode of Life Data System (BOLD) with reference codes linked to each photo. Plotosus lineatus Lineages I–IX colours follow Figs. 2 and 3. For details see ESM Table S1
The Plotosidae clade is further divided into two highly supported clades; one containing all the P. lineatus, P. japonicus and P. limbatus sequences, and the other all the remaining available species of Plotosus (Fig. 1). Plotosus lineatus sequences cluster into nine distinct highly supported lineages, here named Lineages I–IX (Fig. 1). The genetic distances among the nine P. lineatus Lineages ranged from 2% to 16% (Table 1). The highest genetic diversity was seen within Lineage VI (Table 1). The minimum genetic distance observed between P. japonicus and P. lineatus Lineages I–III was 3% while a maximum of 14% was observed between P. japonicus and P. lineatus Lineage IX (Table 1).
Sister to P. lineatus Lineages is a clade containing all P. canius and P. nkunga individuals whose first split corresponds to one highly divergent individual, Plotosus sp., with a minimum of 14% to P. nkunga (Fig. 1; Table 1; ESM Table S1). A unique assembly with the majority of the P. nkunga individuals is sister to five P. canius lineages (P. canius Lineages I–V; Fig. 2; ESM Table S1) and a group with high within-genetic diversity that included several individuals labelled as both species (Fig. 2; ESM Table S1).
Haplotype (TCS) network showing the relationships of all published Plotosus lineatus sequences (ESM Table S2). Circle size is proportional to the observed haplotype frequencies; numbers indicate nucleotide substitutions. Mean genetic divergence (COI) between Lineages (dash lines) is indicated in grey (see Table 1 for details). Plotosus lineatus Lineages I–IX colour scheme follows Figs. 1 and 3. n = Number of individuals; h = haplotypes
All the available individual photos in BOLD were downloaded and linked to each Lineage in the phylogeny (Fig. 1).
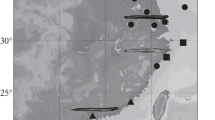
Plotosus lineatus genetic diversity and phylogeographic patterns. The resulting haplotype network allows a more detailed display of spatial patterns according to the distribution of haplotypes (Fig. 2) obtained from the 133 samples of the nine P. lineatus Lineages depicted in the phylogeny. A total of 31 distinct haplotypes were recovered, 15 of which occurred only once with the most frequent haplotype (H5) occurring in 43 individuals across a wide geographical range (ESM Table S2). The geographic mapping of P. lineatus lineages is presented in Fig. 3. We were only unable to link one haplotype (H28; ESM Table S2) with its sampling site. Plotosus lineatus Lineage I displayed a star-like topology, with one high-frequency central haplotype (H5) and several additional haplotypes connected by few-step mutations (Fig. 2). It has the highest number of haplotypes (n = 12) sampled across the widest geographical area (Fig. 3). Haplotype 11 was present in India and also in the Red Sea and the Gulf of Aqaba (Figs. 2, 3; ESM Table S2). Plotosus japonicus single haplotype was placed in an intermediate position, with a minimum of 11 mutations to P. lineatus Lineage I and a minimum of 10 mutations to P. lineatus Lineage II (Fig. 2). Plotosus lineatus Lineage I was further linked by a minimum of 25 mutations to P. limbatus (Fig. 2). Plotosus lineatus Lineages III, IV, VII, VIII and IX were represented by a single haplotype (and are all restricted to a single location each; Figs. 2, 3). Plotosus lineatus Lineage V was only found in the Mediterranean with three haplotypes (Figs. 2, 3; ESM Table S2).
Discussion
Here, we demonstrate that Plotosus spp., and in particular P. lineatus (and P. canius) represent a ‘classic’ case of marine fish cryptic diversity. Cryptic diversity refers to the occurrence of numerous genetically separate populations or species that are morphologically similar (Walker 1964; Struck et al. 2018 supplemental table S4 online for a list of definitions). These populations or species may have diverged from a common ancestor as a result of geographic isolation, reproductive strategies, limited dispersal, habitat adaptation, convergent evolution, or environmental diversity, and may now exhibit distinct behaviours, physiological adaptations, or ecological roles (Fišer et al. 2018). Several species examined in this study share similar morphological features; therefore, they may be mistakenly classified as a single species or lumped into a larger taxonomic group, as is demonstrated here to be the case of striped eel catfish P. lineatus. This may help explain the reason why, despite its long history, the taxonomic status of the P. lineatus species complex remains understudied and uncertain. However, we have uncovered several issues that add complexity to the taxonomic confusion of P. lineatus and therefore, to avoid perpetuating this concern, we have compiled all available information for Plotosus spp. for future use (ESM Table S1).
Lack of adequate data on Plotosus. In our dataset, several incidents of inaccurate data in Plotosus spp. were detected. In some cases, sequences with both BOLD and NCBI entries had different associated manuscripts (e.g., EF607324.1 and EF607323.1 attributed to Zhang 2011 whereas the BOLD IDs, FSCS284-06 and FSCS285-06, are linked to Zhang and Hanner 2012). Others, where the accession number provided has different descriptions (e.g., MF601472 in NCBI is attributed to P. canius whereas in the associated manuscript, Habib et al. 2017, the same accession number is associated with Lepturacanthus savala). In the end, some informed assumptions were made, based on our extensive revision. For example, we have linked sequences EU148553, EU148554 and FJ918911 to the Lakra et al. (2011) information provided in NCBI, although we were unable to find these sequences or any other sequence in the manuscript.
Further aspects add to P. lineatus taxonomic confusion, one being the lack of the exact sampling collection site information. Based on an exhaustive literature review, we detected several situations where this information was missing, i.e., not linked to the analysed individual, neither to the sequence deposited (in either database) nor to the morphological data (e.g., species descriptions). In the majority of the cases, some “wider geographic” information is provided (e.g., Country or Province), although this information is not always directly linked to the data making its discovery challenging. Sometimes, this information is unpublished, others resulted from mistakes [e.g., sequences JF494186.1–JF494188.1 from South Africa in Steinke et al. (2016), but erroneously labelled as being from Lebanon, Mediterranean, in Kundu et al. (2019)]; and sometimes is not available at all (e.g., FJ918911, MZ329579, MW649911 and MW649912). Therefore, we linked all P. lineatus sequences used to their collection locality (ESM Tables S1, S2). We were able to obtain a phylogeographic insight into the nine P. lineatus Lineages (I–IX) for the first time as a result of this valuable data which is covered in more detail below.
Morphology-based misidentifications, sequence similarity search and Plotosus lineatus phylogeny. Lack of adequate data will lead the BLAST searches to ambiguous results. Misidentification of the voucher specimen, contamination during sample processing, inadequate taxonomic identification, or issues with synonyms and syntax could all contribute to this issue. (e.g., Tautz et al. 2003). Morphological confusion between P. lineatus and other (described) species has been reported, e.g., P. lineatus has been mistaken for P. limbatus, frequently seen as the adult of the smaller, striped eel catfish, because both have relatively large eyes and short barbells (Gomon and Taylor 1982). Thus, it is not surprising that our finding of several sequences in both databases (NCBI and BOLD) resulted from morphology-based misidentifications. This problem can mislead the identification of new samples in both databases, due to the sequence similarity search and although previously discussed (e.g., Kundu et al. 2019) has been maintained.
Thus, to avoid the perpetuation of this concern, we compiled all available information for Plotosus spp. for future use (ESM Table S1). The two highly distinct sequences that cluster together in our phylogeny (Unknown species UKFBJ138-08 and KF268174.1 in Fig. 1; ESM Table S1) deposited on NCBI/BOLD as P. lineatus are a clear result of morphology-based misidentifications. On the other hand, the phylogenetic position of the other sequence deposited as Plotosus sp. (MZ606227; Fig. 1; ESM Table S1) does fall within the Plotosidae Clade but is very distantly related to the existent Plotosus sp. sequences available, not allowing for now to draw further comments.
Regarding P. lineatus, we have also found one sequence that was deposited as P. lineatus in both databases (JF952819.1/ABFJ031-06; Fig. 2; ESM Tables S1, S2), even though no morphology-based identification was provided (Zhang and Hanner 2011). This sequence is the same P. japonicus haplotype that was seen in our haplotype network. Another sequence, deposited as P. canius (MN747967.2, unpublished; ESM Table S1) corresponds to haplotype H5 of P. lineatus Lineage I (Fig. 2; ESM Table S2). Finally, three sequences deposited as Plotosus sp. in both databases, correspond to haplotype H30 of P. lineatus Lineage III (Fig. 2; ESM Tables S1, S2). This careful analysis and compilation have also allowed us to increase the number of COI-validated P. lineatus sequences.
Accurate species identification is essential for understanding the nature and extent of marine fish cryptic diversity. By accurately identifying and distinguishing between closely related but genetically distinct species, researchers can improve conservation efforts, understand ecological relationships, and gain insights into the evolutionary history of marine fish communities (e.g., see Ward et al. 2009, for a review). The effectiveness of the barcoding system has been repeatedly demonstrated by the identification of marine fish species with a 2% difference in the DNA barcode as a divergence threshold and a standard cut-off value for species delimitation in fishes (Mabragaña et al. 2011). The present COI Plotosus spp. phylogeny reveals the extent of the hidden complexity of cryptic diversity in species of Plotosus, e.g., nine P. lineatus lineages, ranging from 2% to 15% (Table 1) with two species, P. japonicus and P. limbatus clustering in between them; five P. canius lineages and one group of both species mixed (Fig. 1). The phylogenetic relationships among the nine revealed P. lineatus lineages are generally well established, except for the position of P. lineatus Lineage VII and P. limbatus. This is most likely due to missing taxa/genetic information, and while we analysed 132 P. lineatus spp. individuals, P. lineatus Lineage VII only has one sequence available. Additionally, two other P. lineatus Lineages, i.e., P. lineatus Lineages IV and VIII are also only composed of one sequence each and five P. lineatus Lineages are confined to single geographic areas (Figs. 2, 3; ESM Table S2).
Altogether, our phylogenetic results highlight the crucial need and the urgency to sequence all species of Plotosus described since at the moment only a small fraction of these have molecular data. There is a clear need to increase the sampling within all the identified P. lineatus Lineages to clarify their genetic diversity, followed by in-depth taxonomic analysis, which might allow revisions to describe them as species.
Unravelling the complexity of cryptic diversity and phylogeographic patterns of Plotosus lineatus. Plotosus lineatus was described from the East Indian Ocean in the Indo-Pacific, a region known to be a hotspot of marine fish cryptic diversity, with numerous examples of genetically distinct populations or species that are difficult to distinguish based on their external appearance (e.g., Lutjanus kasmira complex, Miller and Cribb 2007; surgeonfishes (Acanthuridae), DiBattista et al. 2016). The assessment of genetic diversity within P. lineatus has been previously attempted (e.g., Bariche et al. 2015; Kundu et al. 2019; Goren et al. 2020). However, without including individuals from across P. lineatus wide distributional range (natural and introduced/invasive) and using all available mtDNA COI sequences linked to their geographic sampling locations, those results were inconclusive. Here, the phylogeographic patterns of all P. lineatus lineages are provided (Figs. 2, 3). Only P. lineatus and P. canius are known to have cosmopolitan distributions, while the other species have narrow distribution ranges. However, in this study, we show that within P. lineatus, several monophyletic and highly divergent lineages seem to be restricted to small ranges, e.g., P. lineatus Lineages VII, VIII and VIX (Fig. 3; ESM Table S1). Some caution is needed when extrapolating these findings since P. lineatus Lineages VII and VIII are only represented by single sequences and, although the haplotype of P. lineatus Lineage IX has been retrieved in six individuals, they were all, very likely, caught at very close locations (ESM Table S2).
Nevertheless, clear and interesting phylogeographic patterns can be observed. Plotosus lineatus Lineage I is the most geographically widespread, with the highest number of haplotypes and individuals sequenced (Figs. 2, 3; ESM Table S2). This lineage exhibits a star-like topology, with one high-frequency central haplotype and several additional haplotypes connected by few-step mutations, a pattern of connectivity (between haplotypes) that provides support for a recent population expansion. Two haplotypes are geographically widespread; H5, the highly frequent central haplotype, that is present from Indonesia in the East, to India in the West; and H11, sequenced in only seven individuals but present from India in the East, to the Red Sea in the West (Fig. 3; ESM Table S2). Some private haplotypes are also found in P. lineatus Lineage I, e.g., H31 is present only in Seychelles (Fig. 3; ESM Table S2). Plotosus lineatus Lineage II is formed by two haplotypes present in Eastern South Africa, Lineage III seems to be restricted to Madagascar, and Lineage IV is composed of a single sequence from the Réunion Island (Fig. 3; ESM Table S2).
Plotosus lineatus Lineage V is the most puzzling. The geographic mapping of P. lineatus lineages (Fig. 3) reveals that P. lineatus Lineage I is present in the Red Sea and Gulf of Aqaba, so one might expect this lineage to be present in the Mediterranean basin. However, only three haplotypes (retrieved from 29 individuals) of P. lineatus Lineage V are present in the Mediterranean Basin. The first record of P. lineatus in the Mediterranean was in 2002, in Israel (Golani 2002), and since then the species has been recorded in several other coastal locations of the basin, i.e., Lebanon, Syria, Turkey, Egypt, Tunisia and Iran (Bitar 2013; Temraz and Souissi 2013; Ali et al. 2015; Ketabi and Jamily 2016; Ounifi-Ben Amor et al. 2016; Bayhan and Ergüden 2022). Unfortunately, none of these records includes molecular data. Our results reveal P. lineatus Lineage V with a minimum genetic distance of 4% to Lineage VI and a maximum of 13% to Lineage IX (Table 1). Lineage VI is present from Vietnam in the East, to Saudi Arabia Gulf in the West, with no records found in the Red Sea (Fig. 3; ESM Table S2). Several hypotheses could explain the exclusive presence of P. lineatus Lineage V in the Mediterranean. First, this lineage could be endemic in the Mediterranean. This hypothesis seems unlikely because striped eel-catfish is a very charismatic species, morphologically very easy to identify and it has only been recorded for the first time in the Mediterranean in 2002. A more likely explanation would be that P. lineatus Lineage V migrated from the Red Sea to the Mediterranean Sea, through the Suez Canal, a pattern described for several other fish species (Bentur et al. 2018). However, no other haplotypes were found outside the Mediterranean and this lineage has a minimum genetic distance of 9% to Lineage I (Table 1), the only lineage found in the Red Sea so far (Fig. 3). There should be yet, other hypotheses to consider. For example, the presence of P. lineatus Lineage V in the Mediterranean could result from the release of striped eel catfish individuals from aquarium-imported and/or ship-ballast water, whose natural geographic range has not yet been sampled. This species is commercially important for both the aquarium trade and human consumption (Situ and Sadovy 2004; Scandol and Rowling 2007; Asriyana et al. 2021, 2022) and the “escaped/released” individuals would have been able to survive and establish in the Mediterranean. However, an important question remains unanswered: is this P. lineatus Lineage V the one that has been continuously recorded in the Mediterranean since 2002? Is it the genetic pool that has been assessed as a high-risk marine invasive alien species for the EU and included in the list of invasive alien species of EU concern? Or is there another P. lineatus lineage, most likely P. lineatus Lineage I, already present in the Red Sea, in the Mediterranean? There is, therefore, an urgent need to investigate the complete molecular diversity of Plotosus spp. in the Mediterranean.
Finally, our extensive compilation and haplotype geographic mapping revealed the existence of several pairs of species/Lineages living in sympatry, e.g., P. limbatus and P. lineatus Lineage I, P. lineatus Lineage I and P. lineatus Lineage VI (at two different locations), P. nkunga and P. canius, P. canius and P. lineatus Lineage I (Fig. 3; ESM Tables S1, S2). These findings raise an array of additional questions, which remain unanswered for the time being.
Final remarks. Overall, the Plotosus genus, and P. lineatus in particular, represents a fascinating example of marine fish cryptic diversity in the Indo-Pacific region, with numerous genetically distinct but morphologically similar species that are extremely difficult to distinguish based on external characteristics alone. The current study identified numerous significant roadblocks to the clarification of P. lineatus taxonomy and revealed the existence of nine P. lineatus Lineages for the first time, along with their known geographic range. We highlighted the urgent need for taxonomic revisions to describe them as species.
References
Ali M, Abdel-Baki AAS, Abdel-Ghaffar F (2007) Zschokkella egyptica n. sp. (Myxosporea: Bivalvulaida) infecting the gallbladder of the eel catfish Plotosus lineatus Thunberg, 1787 and the freckled goatfish Upeneus tragula Richardson, 1846 in the Red Sea, Egypt. Parasitol Res 100:625–628
Ali M, Saad A, Soliman A (2015) Expansion confirmation of the Indo-Pacific catfish, Plotosus lineatus (Thunberg, 1787), (Siluriformes: Plotosidae) into Syrian marine waters. Am J Biol Life Sci 3:7–11
Asriyana A, Halili H, Balubi AM, Asrari A, Kurnia A (2022) Growth performance and survival rate of striped eel catfish (Plotosus lineatus) fingerlings in different microhabitats. Aquac Aquar Conserv Legis 15:2775–2784
Asriyana A, Halili H, Hamzah M, Kurnia A (2021) Growth performance and survival rate of striped eel catfish (Plotosus lineatus) in the domestication. Biodiversitas 22:5593–5599
Bariche M, Torres M, Smith C, Sayar N, Azzurro E, Baker R, Bernardi G (2015) Red Sea fishes in the Mediterranean Sea: a preliminary investigation of a biological invasion using DNA barcoding. J Biogeogr 42:2363–2373
Bayhan YK, Ergüden D (2022) Occurrence of Plotosus lineatus (Thunberg, 1787) from the northeastern Mediterranean coast of Turkey. Mar Life Sci 4:142–145
Bentur Y, Altunin S, Levdov I, Golani D, Spanier E, Edelist D, Lurie Y (2018) The clinical effects of the venomous Lessepsian migrant fish Plotosus lineatus (Thunberg, 1787) in the southeastern Mediterranean Sea. Clin Toxicol (Phila) 56:327–331
Betancur-R R, Wiley EO, Arratia G, Acero A, Bailly N, Miya M, Lecointre G, Orti G (2017) Phylogenetic classification of bony fishes. BMC Evol Biol 17:162
Bitar G (2013) Sur la présence des poissons exotiques nouveaux de la côte libanaise (Méditerranée orientale). Rapp Comm Int Mer Médit 40:592
Boulenger GA (1895). Descriptions of two new fishes obtained by Mr. C. Hose in Sarawak. Annal Mag Natl Hist (Ser 6) 15 (87) (31): 247
Burgess WE (1989) An atlas of freshwater and marine catfishes. A preliminary survey of the Siluriformes.T.F.H. Publications, Neptune City
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973
Clement M, Posada DC, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Cuvier G, Valenciennes A (1840). Histoire naturelle des poissons. Tome quinzième. Suite du livre dix-septième. Siluroïdes. F G Levrault, Paris
DiBattista JD, Whitney J, Craig MT, Hobbs JPA, Rocha LA, Feldheim KA, Berumen ML, Bowen BW (2016) Surgeons and suture zones: hybridization among four surgeonfish species in the Indo-Pacific with variable evolutionary outcomes. Mol Phylogenet Evol 101:203–215
Dogdu SA, Uyan A, Uygur N, Gürlek M, Ergüden D, Turan C (2016) First record of the Indo-Pacific striped eel catfish, Plotosus lineatus (Thunberg, 1787) from Turkish marine waters. Nat Eng Sci 1(2):25–32
Edelist D, Golani D, Rilov G, Spanier E (2012) The invasive venomous striped eel catfish Plotosus lineatus in the levant: possible mechanisms facilitating its rapid invasional success. Mar Biol 159:283–290
Fahim FA, Mady EA, Ahmed SM, Zaki MA (1996) Biochemical studies on the effect of Plotosus lineatus crude venom (in vivo) and its effect on EAC-cells (in vitro). Adv Exp Med Biol 391:343–355
Ferraris C (1999) Plotosidae: eeltail catfishes (also, eel catfishes, stinging catfishes, and coral catfishes). In: Carpenter KE, Niem VH (eds) FAO species identification guide for fishery purposes. The living marine resources of the Western Central Pacific, vol. 3. Batoid fishes, chimaeras and Bony fishes, part 1 (Elopidae to Linophrynidae). FAO, Rome, pp 1880–1883
Fišer C, Robinson CT, Malard F (2018) Cryptic species as a window into the paradigm shift of the species concept. Mol Ecol 27:613–635
Fricke, R. & Eschmeyer, W. N. 2023. Guide to Fish Collections. Electronic version. http://researcharchive.calacademy.org/research/ichthyology/catalog/collections.asp. Accessed January 2023
Froese R, Pauly D (2023) FishBase. Electronic version. http://www.fishbase.org. Accessed January 2023
Galanidi M, Turan C, Öztürk B, Zenetos A (2019) Europen Union (EU) risk assessment of Plotosus lineatus (Thunberg, 1787); a summary and information update. J Black Sea Mediterr Environ 25:210–231
Galanidi M, Zenetos A, Bacher S (2018) Assessing the socio-economic impacts of priority marine invasive fishes in the Mediterranean with the newly proposed SEICAT methodology. Mediterr Mar Sci 19:107–123
Golani D (2002) The Indo-Pacific striped eel catfish, Plotosus lineatus (Thunberg, 1787) (Osteichtyes: Siluriformes), a new record from the Mediterranean. Sci Mar 66:321–323
Gomon JR, Taylor WR (1982) Plotosus nkunga, a new species of catfish from South Africa, with a redescription of Plotosus limbatus Valenciennes and a key to the species of Plotosus (Siluriformes: Plotosidae). J L B Smith Inst Ichthyol Special Publ 22:1–16
Goren M, Stern N, Diamant A (2020) New records of Plotosus japonicus in the Red Sea and genetic indications for its presence throughout the Indo-Pacific (Osteichthyes: Plotosidae). Zool Middle East 66:124–131
Habib KA, Kim CG, Oh J, Neogi AK, Lee YH (2017) Aquatic biodiversity of Sundarbans, Bangladesh (2nd edition). Korea Institute of Ocean Science and Technology (KIOST), Busan Metropolitan City
Hamilton F (1822). An account of the fishes found in the river Ganges and its branches. Edinburgh, London
Hebert PD, Cywinska A, Ball SL, DeWaard JR (2003) Biological identifications through DNA barcodes. Proc R Soc B Biol Sci 270:313–321
Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Ketabi R, Jamily S (2016) Plotosus lineatus (Thunberg, 1787). Iranian Fisheries Science Research Institute, Islamic Azad University, Science and Research Branch, Tehran
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol 35:1547–1549
Kundu S, Pakrashi A, Laskar BA, Rahaman I, Tyagi K, Kumar V, Chandra K (2019) DNA barcoding reveals distinct population of Plotosus canius (Siluriformes: Plotosidae) in Sundarbans waters. Mitochondrial DNA B Resour 4:1167–1171
Lacepède BGE (1803). Histoire naturelle des poissons. Tome Cinquieme 5: i–lxviii + 1–803
Lakra WS, Verma MS, Goswami M, Lal KK, Mohindra V, Punia P, Gopalakrishnan A, Singh KV, Ward RD, Hebert P (2011) DNA barcoding Indian marine fishes. Mol Ecol Resour 11:60–71
Lanzing W (1967) A possible relation between the occurrence of a dendritic organ and the distribution of the Plotosidae (Cypriniformes). Pac Sci 21:498–502
Leigh JW, Bryant D (2015) POPART: full-feature software for haplotype network construction. Methods Ecol Evol 6:1110–1116
Mabragaña E, de Astarloa JMD, Hanner R, Zhang J, Castro MG (2011) DNA barcoding identifies Argentine fishes from marine and brackish waters. PLoS One 6(12):e28655
Malakpour Kolbadinezhad S, Coimbra J, Wilson JM (2018a) Effect of dendritic organ ligation on striped eel catfishPlotosus lineatus osmoregulation. PLoS One 13(10):e0206206
Malakpour Kolbadinezhad S, Coimbra J, Wilson JM (2018b) Osmoregulation in the Plotosidae catfish: Role of the salt secreting dendritic organ. Front Physiol 9:761
Malakpour Kolbadinezhad S, Coimbra J, Wilson JM (2020) Is the dendritic organ of the striped eel catfish Plotosus lineatus an ammonia excretory organ? Comp Biochem Physiol A Mol Integr Physiol 241:110640
Miller TL, Cribb TH (2007) Phylogenetic relationships of some common Indo-Pacific snappers (Perciformes: Lutjanidae) based on mitochondrial DNA sequences, with comments on the taxonomic position of the Caesioninae. Mol Phylogenet Evol 44:450–460
Myers RF (1999) Micronesian reef fishes. A comprehensive guide to the coral reef fishes of Micronesia. Coral Graphics. Barrigada, Guam
Ng HH, Sparks JS (2002) Plotosus fisadoha, a new species of marine catfish (Teleostei: Siluriformes: Plotosidae) from Madagascar. Proc Biol Soc Wash 115:564–569
Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274
Ounifi-Ben Amor K, Rif M, Ghanem R, Draief I, Zaouali J, Souissi JB (2016) Update of alien fauna and new records from Tunisian marine waters. Mediterr Mar Sci 17:124–143
Prokofiev A (2008). A new species of eel catfishes of the genus Plotosus Nha Trang Bay (South China Sea, central Vietnam) (Siluriformes: Plotosidae). J Ichthyol 48:671–675
Scandol J, Rowling K (2007) Resource assessments for multi-species fisheries in NSW, Australia: qualitative status determination using life history characteristics, empirical indicators and expert review. International Council for the Exploration of the Sea, Helsinki
Shao F, Wu ZC, Xu Y, Li P, Peng ZG (2023) Genome of striped eel catfish (Plotosus lineatus) provides insights into the adaptive evolution of amphidromous fish. Zool Res 44:349–352
Shiomi K, Takamiya M, Yamanaka H, Kikuchi T, Suzuki Y (1988) Toxins in the skin secretion of the oriental catfish (Plotosus lineatus): immunological properties and immunocytochemical identification of producing cells. Toxicon 26:353–361
Situ YY, Sadovy YJ (2004) A preliminary study on local species diversity and seasonal composition in a Hong Kong wet market. Asian Fish Sci 17:235–248
Steinke D, Connell AD, Hebert PD (2016) Linking adults and immatures of South African marine fishes. Genome 59:959–967
Streftaris N, Zenetos A (2006) Alien marine species in the Mediterranean—the 100 ‘worst invasives’ and their impact. Med Mar Sci 7(1):87–118
Struck TH, Feder JL, Bendiksby M, Birkeland S, Cerca J, Gusarov VI, Kistenich S, Larsson H, Liow LH, Nowak MD, Stedje B, Bachmann L, Dimitrov D (2018) Finding evolutionary processes hidden in cryptic species. Trends Ecol Evol 33:153–163
Tautz D, Arctander P, Minelli A, Thomas RH, Vogler AP (2003) A plea for DNA taxonomy. Trends Ecol Evol 18:70–74
Taylor WR, Gomon JR (1986) Plotosidae. In: Daget J, Gosse J-P, Thys van den Audenaerde DFE (eds) Check-list of the freshwater fishes of Africa. Vol. 2. ISNB, Bruxelles, MRAC, Tervuren, ORSTOM, París, pp 160–162
Temraz T, Souissi JB (2013) First record of striped eel catfish Plotosus lineatus (Thunberg, 1787) from Egyptian waters of the Mediterranean. Rapp Comm Int Mer Medit 40:604
Thunberg CP (1787) Museum naturalium Academiae Upsaliensis...Praeside C.P. Thunberg, etc., Part 1. Upsaliae, Mus Acad, Upsaliensis
Wakwabi E, Mees J (1999) The epibenthos of the backwaters of a tropical mangrove creek (Tudor Creek, Mombasa, Kenya). Neth J Zool 49:189–206
Walker TJ (1964) Cryptic species among sound-producing ensiferan Orthoptera (Gryllidae and Tettigoniidae). Q Rev Biol 39:345–355
Ward RD, Hanner R, Hebert PD (2009) The campaign to DNA barcode all fishes, FISH‐BOL. J Fish Biol 74:329–356
Weber M (1910). Neue Fische aus Niederländisch Süd-Neu-Guinea. Note Leyden Mus 32 (4) (28): 225–240
Yoshino T, Kishimoto H (2008). Plotosus japonicus, a new eeltail catfish (Siluriformes: Plotosidae) from Japan. Bull. Natl Mus Natu Sci (Ser A) Suppl 2: 1–11
Zhang J (2011). Species identification of marine fishes in China with DNA barcoding. Evidence-Based Complementary and Alternative Medicine 2011:978253
Zhang JB, Hanner R (2011) DNA barcoding is a useful tool for the identification of marine fishes from Japan. Biochem Syst Ecol 39:31–42
Zhang J, Hanner R (2012) Molecular approach to the identification of fish in the South China Sea. PLoS One 7(2):e30621
Acknowledgements
This study was funded by ATLANTIDA - Platform for the monitoring of the North Atlantic Ocean and tools for the sustainable exploitation of the marine resources (NORTE-01-0145-FEDER-000040). AGS was funded under the Grant 2023_033_BI_ATLANTIDA. EF is supported by FCT—Foundation for Science and Technology (contract CEECINST/00027/2021). We thank the anonymous reviewers and the editor for the helpful remarks and suggestions, which have improved the manuscript.
Funding
Open access funding provided by FCT|FCCN (b-on).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
N/A.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10228_2023_924_MOESM1_ESM.treefile
Supplementary file1 ESM File S1 Phylogenetic tree in newick format used to construct the phylogeny presented in Fig. 1 (TREEFILE 17 KB)
10228_2023_924_MOESM2_ESM.xlsx
Supplementary file2 ESM Table S1 Detailed information associated with all the Plotosus spp. sequences used in this study, retrieved from the Nucleotide Center for Biotechnology Information (NCBI) and Barcode of Life Data System (BOLD). Haplotype numbers and Plotosus spp. Lineages refer to the text (XLSX 107 KB)
10228_2023_924_MOESM3_ESM.xlsx
Supplementary file3 ESM Table S2 Detailed information associated with the Plotosus lineatus Lineages I–IX sequences used to construct the haplotype network (XLSX 42 KB)
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Froufe, E., Gomes-dos-Santos, A., Matos, A. et al. How complex is the hidden species diversity of the teleost Plotosus genus?. Ichthyol Res 71, 163–173 (2024). https://doi.org/10.1007/s10228-023-00924-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10228-023-00924-2