Abstract
We analyzed two morpho-species, Astyanax aeneus and Astyanax caballeroi, coexisting in Lake Catemaco, with ecological differences previously associated with different trophic niches. While A. aeneus is a widely distributed species, A. caballeroi is endemic to Lake Catemaco. Due to the contrasting morphology between these two sympatric species, they were originally assigned to different genera, but little is known about their genetic differentiation. We hypothesized that these two species, which present differentiation in morphology and in trophic niches, co-occur in concomitance with assortative mating and form distinct genetic clusters. We tested this prediction by typing a set of 12 microsatellites in 348 individuals of the two morpho-species, comparing their genetic structure patterns with that of an allopatric population of A. aeneus (i.e., the Maquinas population). Genetic structure analysis, assignment analyses, and estimation of gene flow between the sympatric morpho-species were carried out. Contrary to expectations, there is a lack of genetic differentiation between the A. aeneus and A. caballeroi morpho-species. However, there was a genetic differentiation between Lake Catemaco and Río Maquinas populations of A. aeneus. In addition, we found an asymmetric gene flow pattern, with a larger migration rate from A. aeneus to A. caballeroi than in the other direction. Finally, the allopatric population of A. aeneus showed lower levels of genetic diversity than those reported for both morpho-species in the lacustrine system. Consequently, our results support the notion that these morpho-species are either in an early phase of speciation or represent a single polymorphic species. This model provides relevant information to understanding the presence of polymorphisms under gene flow and their potential impact on the speciation process.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The origin and maintenance of biodiversity remain challenging areas of research in biology. Ecological divergence has been recognized as a force promoting diversity and speciation (Schluter 1996). Under this scenario, adaptive evolution related to different ecological conditions promotes the emergence of alternative phenotypes that could differ in fitness and cause assortative mating (West-Eberhard 2003; Dieckmann et al. 2004; Nosil 2012; Bay et al. 2017).
Ecological speciation occurs when adaptation to different environments or resources causes the evolution of irreversible reproductive barriers (Doebeli and Dieckmann 2003), but in many cases, the adaptive divergence is associated with weak to modest levels of reproductive isolation (Thibert-Plante and Hendry 2009; Kautt et al. 2016a). In this regard, ecology-driven speciation is thought of as a gradual process in which different measures of divergence can be used to assess the ‘stage’ of speciation (Nosil et al. 2009). Without necessarily being in order such stages generally include the emergence of phenotypic, ecological, behavioral, and genetic divergence. However, at the later stages of the speciation process, we would expect a complete genetic divergence (see Doebeli and Dieckmann 2003; Nosil 2012) corresponding to the two ecologically divergent lineages. Thus, during the speciation process, the reproductive isolation could range from absent to complete, as in the examples of Timema walking-stick insects (absent, Nosil et al. 2009) to some rare examples such as a pair of sympatric Amphilophus cichlids in Lake Apoyo (complete, Barluenga et al. 2006; Kautt et al. 2020).
In the geographic context, ecological speciation can occur independently of whether reproductive isolation evolves in sympatry, parapatry, or allopatry (Hatfield and Schluter 1999; Schliewen et al. 2001; Bolnick et al. 2009; Bolnick 2011). However, in sympatry, we can expect different levels of genetic isolation depending on how these phenotypic differences affect their fitness and assortative mating. In this regard, recent studies have suggested that the genetic architecture of divergent traits can influence whether sympatric species occur (Kautt et al. 2020). Simple monogenic or oligogenic traits may show only localized differentiation, in contrast to polygenic traits, which can lead to genome-wide differentiation and thus promote barriers to gene flow that ultimately trigger speciation (Kautt et al. 2020).
The Astyanax genus is characterized by its ability to adapt to a wide range of ecological conditions and is considered a model system in the study of phenotypic evolution (Jeffery 2009; McGaugh et al. 2020). The genus includes some striking examples of morphological divergence associated with extreme habitats, such as those reported in the cavefish Astyanax mexicanus (e.g., Protas et al. 2007; Gross et al. 2009; Jeffery 2009; Elipot et al. 2014; McGaugh et al. 2014).
Another extraordinary example of highly divergent morphs corresponds to the lake-dwelling fishes distributed in lacustrine systems across Mexico and Central America, originally recognized as different genera (i.e., Astyanax and Bramocharax) (see Powers et al. 2020). Previous phylogenetic studies based on both morphological and molecular data recovered the Bramocharax genus as polyphyletic (Ornelas-García et al. 2008; Schmitter-Soto 2017), suggesting a parallel evolution of lacustrine divergent pairs of ecotypes within the Astyanax genus. These lacustrine divergent pairs are distributed in each of the two lakes of Mexico (i.e., Lake Catemaco and Lake Ocotalito), and in different hydrological systems of Central America (e.g., in Guatemala, Lake Managua and Lake Nicaragua, in Nicaragua and Costa Rica) (Rosen 1972; Contreras-Balderas and Rivera-Teillery 1983; Ornelas-García et al. 2008; Powers et al. 2020; Garita-Alvarado et al. 2018, 2021). Interestingly, these divergent ecotypes show a clinal variation in the craniofacial morphology and dentition, with the largest differentiation between the Central American sympatric morphs, and the lowest divergence in the Mexican lacustrine populations (Powers et al. 2020). However, even when the levels of disparity in the Mexican systems were lower, we observed that both craniofacial and trophic trait divergence was in the same direction as in the Central American systems; that is, the morphs differ in their jaw elongation and width, skull depth, and in the number of maxillary teeth and tooth size (Powers et al. 2020). Despite our expectations, when we assessed the genetic differentiation between the Central American ecotypes, they showed a marginal genetic differentiation based on microsatellite data, suggesting an incipient speciation process or tentatively a stable polymorphism (Garita-Alvarado et al. 2021), raising questions about the levels of genetic differentiation in the other lacustrine systems.
In the present study, we focus on one lacustrine system of Mexico, Lake Catemaco, where the two morpho-species (i.e., diagnosed based on morphological differences) Astyanax aeneus and Astyanax caballeroi coexist (Fig. 1) and are currently recognized as valid species (Schmitter-Soto 2017). In previous studies, we documented their phenotypic and ecomorphological divergence (Ornelas-García et al. 2014, 2018). Even more so, considering the strong morphological differences between these two sympatric morpho-species, mainly in trophic related traits (Ornelas-García et al. 2018), taxonomists originally considered them as distinct genera, with A. caballeroi being originally described within the genus Bramocharax, whose diagnostic features were the shape of the teeth and the dentary formula in the different oral bones (i.e., the number of maxillary and dentary teeth). However, nowadays, there is a taxonomic consensus that they correspond to the same genus, Astyanax, but to different species (Schmitter-Soto 2017). Among the diagnostic traits that differentiate A. caballeroi from A. aeneus are a larger mean dentary tooth length, a higher number of maxillary teeth, larger eye size, a longer snout, a slender body, and a concave head profile with an upward mouth orientation, in contrast to its A. aeneus counterpart (Contreras-Balderas and Rivera-Teillery 1983; Ornelas-García et al. 2008, 2014; Powers et al. 2020). Recently, we corroborated that the ecomorphological differentiation between the two morpho-species is associated with their trophic habits, with A. caballeroi showing a higher value of δ15N (Ornelas-García et al. 2018). Moreover, the differences in the stable isotope values were also associated with gut contents, with a 10-fold higher proportion of invertebrates in A. caballeroi than in A. aeneus. This ecomorphological–trophic association provides additional evidence of an ecological niche partitioning between the two currently recognized species (Ornelas-García et al. 2018).
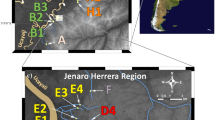
Map of the sampled localities in the Catemaco Lake. a Astyanax caballeroi and b A. aeneus morpho-species, lines below images correspond to 10 mm as a size reference. Circles, sampling points from Catemaco Lake and Maquinas River. 1 Changos Island, 2 Agatepec Island, 3 Finca, 4 Maxacapan, 5 La Victoria, 6 Pozolapan, 7 Catemaco, 8 Ecuniapan, 9 Mimiagua, 10 Las Margaritas, 11 Cuetzalapan, 12 Oxochapan, 13 Maquinas River. The color of the dots coincides with the correspondence factor analysis in Fig. 2
Despite clear evidence of the ecological divergence between these species from Lake Catemaco, little is known about their genetic differentiation. Previous phylogenetic and phylogeographic studies, primarily based on mitochondrial data, have shown no differentiation with shared haplotypes between the two sympatric morpho-species (Ornelas-García et al. 2008, 2014). Thus, in the present study, we evaluate the genetic structure as well as the gene flow between the two sympatric morpho-species, A. aeneus, and A. caballeroi, and with an allopatric population of A. aeneus based on 12 microsatellite loci. Surprisingly, we find no signs of any genetic differentiation within the lake, even with the most sensitive analysis procedures. Therefore, we suggest that this system is at its earliest possible stage of ecological speciation, or may correspond to stable monogenic or oligogenic polymorphisms, where the phenotypic differences could be driven by a small number of loci and phenotypic differentiation has not yet led to genetic differentiation.
Methods
Sample Collection and DNA Extraction. A total of 318 individuals assigned to both nominal species were collected in 2006 at 13 locations in Lake Catemaco, Mexico [Fig. 1; Electronic Supplementary Material (ESM) Table S1]. We used the species names Astyanax aeneus and A. caballeroi, since they were collected according to these original diagnostic traits (Contreras-Balderas and Rivera-Teillery 1983). One allopatric population of A. aeneus from the Maquinas River basin, near Lake Catemaco, was included to test if the microsatellite loci used were able to differentiate between allopatric populations. A small fin clip sample was taken from all individuals and the remainder was preserved in ethanol as a voucher for future morphological analyses. The fin clips were also preserved in 90% ethanol and subsequently stored at -20 °C. DNA was extracted using a standard Proteinase-K in SDS/EDTA digestion and NaCl (4.5 M) and chloroform, as described in Sonnenberg et al. (2007). Vouchers were deposited at the Museo Nacional de Ciencias Naturales, CSIC, Madrid, Spain.
Twelve microsatellite loci were taken from a previous study in a closely related species A. mexicanus (Protas et al. 2006). These loci correspond to dinucleotide-repeat sequences and were chosen considering their location on different chromosomes. The loci were multiplexed in two reactions using a multiplex PCR kit (QIAGEN), in 5 μL reaction final volumes following kit instructions. PCR amplifications consisted of one cycle of denaturation at 95 °C for 5 min; 35 cycles of 94 °C for 30 s, annealing for 30 s at 52–60 °C, and extension at 72 °C for 45 s; then followed by one cycle of 7-min extension at 72 °C (ESM Table S2). Forward primers were labeled with fluorescent dyes (Invitrogen), and amplified PCR products were run on an ABI Prism 3730 DNA Analyzer (GS500 ROX size standard). All loci tested were successfully amplified after PCR optimization. Of those, all were polymorphic and were used in the full screening of all samples (total N = 348; ESM Table S2) following the protocol described previously. Allele scoring was performed using Geneious v. 10.2.3 (Biomatters).
Genetic diversity analyses. Deviations from Hardy–Weinberg (HW) proportions were tested using the exact probability test for multiple alleles (Guo and Thompson 1992) available in Genepop v. 4.0.1 (Raymond 1995) at each locus for each population and overall loci for each population. Genotypic linkage disequilibrium between each pair of loci was estimated by Fisher’s exact tests with Genepop v. 4.0.1. Both tests for deviations from HW proportions and linkage disequilibrium used a Markov chain (10,000 dememorization steps, 1,000 batches, 2,000 iterations per batch) (Guo and Thompson 1992). Correction for multiple testing (type I error rates) was performed using the sequential Bonferroni (Rice 1989). Additionally, MICRO-CHECKER v. 2.23 (Van Oosterhout et al. 2004) was used to explore the existence of null alleles and to evaluate their impact on the estimation of genetic differentiation.
Microsatellite genetic diversity was quantified for locus and sampling site based on the average number of alleles per locus (NA), the number of alleles standardized to those of the population sample with the smallest size [NS (Nei and Chesser 1983)], and the observed (Ho) and expected (He) heterozygosities (Nei 1978) using GENETIX v.4.05.2 (Belkhir 2004) and Fstat 2.9.3 (Goudet 2001).
Population structure and gene flow analyses. To visualize the relationship between population samples, and between A. aeneus and A. caballeroi, a factorial correspondence analysis (Guinand 1996) was implemented in GENETIX v.4.05.2 (Belkhir 2004). Additionally, a Bayesian clustering method was used to assess the possibility of genetic structure between the morpho-species. The number of populations (K) with the highest posterior probability (mean lnProb [D]) was calculated with the program STRUCTURE v. 2.0 (Pritchard et al. 2000), assuming an admixed model and a uniform prior probability of the number of populations, K. MCMC simulations consisted of 5 × 105 burn-in iterations followed by 4.5 × 106 sampled iterations. Furthermore, the modal value of λ, ΔK (Evanno et al. 2005) was calculated to infer the best value of K. Ten runs for each value of K were conducted to check for consistency in the results.
In addition, discriminant analysis of principal components (DACP) (Jombart et al. 2010, 2011) was performed in R v. 4.2.2 (R Core Team 2022), in RStudio v. 1.4. (RStudio Team 2022). DAPC is a multivariate analysis designed to identify and describe clusters of genetically related individuals. DAPC relies on data transformation using principal component analysis (PCA) as a prior step to discriminant analysis (DA), ensuring that variables submitted to DA are uncorrelated. The DA method defines a model in which genetic variation is partitioned into a between-group and a within-group component and yields synthetic variables that maximize the first while minimizing the second (Jombart et al. 2010). DAPC was performed using microsatellite data with two different strategies: 1) the individual assignment considering as prior the two morpho-species (i.e., A. aeneus vs. A. caballeroi) and 2) using the find.clusters function. We removed missing data and then ran a K-means clustering algorithm (which relies on the same model as DA) with different numbers of clusters, each of which gives rise to a statistical model and an associated likelihood. With the find.clusters function, we evaluated from K = 1 to K = 14 possible clustering in 10 different iterations (DAPC) (Jombart et al. 2011). In both strategies, the selection of the number of principal components was carried out with a cross-validation analysis. The validation set is selected with stratified random sampling, which guarantees that at least one member of each conglomerate or cluster is represented in both the training set and the validation set (Jombart and Collins 2015). The clusters or conglomerates resulting from the DAPC were visualized in a scatter diagram, using the first two discriminant functions, representing individuals as points. The proportions of intermixes, obtained from the membership probability based on the retained discriminant functions, were plotted for each individual. The DAPC admixture index was plotted using StructuRly (Criscuolo and Angelini 2020).
The FST pairwise comparisons were carried out between sympatric species from Lake Catemaco, A. aeneus and A. caballeroi, and each of them vs. the allopatric A. aeneus from the Maquinas River. The FST p values were estimated based on 1,000 permutations in Arlequin, v. 3.5. (Excoffier and Lischer 2010). To determine the amount of genetic structuring among grouping levels, an analysis of molecular variance (AMOVA) was performed with Arlequin, v. 3.5. (Excoffier and Lischer 2010). Based on the FST results, we carried out an AMOVA considering the geographic scheme. Thus, we grouped the two sympatric species (i.e., A. aeneus and A. caballeroi in Lake Catemaco) and compared them with their allopatric A. aeneus from the Maquinas River.
MIGRATE 3.2.6 (Beerli and Felsenstein 1999, 2001) was used to infer the population size parameter Θ (where Θ = 4 Neμ; Ne is the effective population size and μ is the mutation rate per site) and the migration rate M (where M = m/μ and m is the immigration rate per generation) for both A. aeneus and A. caballeroi. Due to the presence of two mitochondrial lineages in both morpho-species (see Ornelas-García et al. 2018), the lineage information was also considered in this analysis, resulting in the following configuration: Astyanax aeneus from lineage 1 and 2, and Astyanax caballeroi from lineage 1 and 2. Based on the estimates of historical migration rates (M) obtained, we tested different models of gene flow between species and lineages, using a Bayes factor approach (Beerli and Palczewski 2010). For the two species, we evaluated the following models: 1) panmictic model with one population size, 2) two population sizes and one migration rate from A. aeneus to A. caballeroi, and 3) two population sizes and one migration rate from A. caballeroi to A. aeneus. The models were compared using Bézier thermodynamic integration (Beerli and Palczewski 2010), and their marginal likelihoods were then used to calculate Bayes factors and model probabilities (Kass and Raftery 1995). We assumed a Brownian-motion mutation model with constant mutation across all loci, and FST and a UPGMA tree were the starting parameters for the estimation of Θ and M. MCMC consisted of ten short chain samplings (of 50,000 trees) and three long chain samplings (of 500,000 trees) using an adaptive heating scheme.
Results
All microsatellite loci were polymorphic in all locations and showed different levels of polymorphism across sites. MICRO-CHECKER indicated the presence of null alleles at two loci across sampling localities. Corrected estimates of FST by null allele presence were very similar to non-corrected values; consequently, all loci were used in the analyses. No linkage disequilibrium was observed between any loci pairs, which suggests the independence of all loci (ESM Table S3). A total of 273 alleles were obtained, ranging from 13 (NYU14) to 32 (NYU29) per locus (mean Na = 22.21) (ESM Table S4), and the number of alleles between the sympatric Astyanax aeneus and A. caballeroi was very similar, with a mean of 20.3 for A. aeneus and 18.58 for A. caballeroi. The allopatric A. aeneus from the Maquinas River showed a considerably lower number of alleles (Na = 3.97). The amount of genetic variability was relatively high across loci and morpho-species clusters (HO ranging from 0.847 to 0.855) (Table 1), while the Maquinas River population showed the lowest Ho and He (0.60 and 0.57, respectively).
Lack of genetic structure among sympatric morpho-species. A factorial correspondence analysis segregated the population of Maquinas River from those of Lake Catemaco, but A. aeneus and A. caballeroi were not differentiated (Fig. 2a). The Bayesian clustering analysis with STRUCTURE, including the Maquinas River population, revealed the highest likelihood for the model with K = 2, in congruence with the value for the highest ΔK for K = 2 (Fig. 2b). Under this scenario, two genetic groups were recovered corresponding to the Maquinas River vs. Lake Catemaco. The latter was recovered as one genetic cluster, supporting admixture between the two morpho-species from Lake Catemaco. No additional peaks in the ΔK were evident (Fig. 2c, d).
Genetic structure by Bayesian cluster analysis based on microsatellite data. a Factorial correspondence analysis for 12 microsatellite loci including the population from the Maquinas River and the Lake Catemaco localities (colored dots). b Bayesian population assignment based on 12 microsatellite loci with the software STRUCTURE for sampling locations from Lake Catemaco (10) and the Maquinas River, showing the grouping with the highest ΔK (for K = 2). c Plot of mean likelihood L(K) and variance per K value from STRUCTURE. d We used Evanno’s test based on Delta K plot to detect the K number of groups that best fit the data (Evanno et al. 2005). CAT Center of Catemaco Lake, CHA Isla Changos, CUL Cuetzalapan, CUR Cuetzalapan River, FIS La Finca, AGA Isla Agatepec, MAR Margaritas, MAX Maxacapan, MIM Mimiagua, OJO Oxochapan, POZ Pozolapan, VIC La Victoria, MAQ Maquinas River
In the DAPC, the microsatellite data matrix had 306 individuals, after removing the missing data. Based on our first strategy considering the two morpho-species a priori on the DAPC, we found a lack of differentiation between them, while the allopatric population of Maquinas River was well differentiated (Fig. 3). The cross-validation test resulted in the retention of 45 principal components (PCs) with an accumulative variance of 69.1% of the total data. On the find.clusters algorithm from the 10 runs performed, we recovered K = 4 corresponding to the lowest BIC value (i.e., 506.54). None of these four clusters correspond to any of the sympatric morpho-species within Lake Catemaco (A. aeneus and A. caballeroi), with the Maquinas River population the only cluster recovered as differentiated. The cross-validation test resulted in the retention of 14 PCs with an accumulative variance of 36.4% of the total data. In the scatterplot of individuals on the two main components of DAPC, we can observe the extensive admixture between the sympatric morpho-species, with the Maquinas River population being well differentiated (ESM Fig. S1).
Assignment of population genetic structure by DAPC based on microsatellite data. a Scatterplot of the discriminant analysis of the 16 principal components based on 12 microsatellite data with K = 3 (i.e., Astyanax aeneus, A. caballeroi, and Maquinas = Maq). The axes represent the first two linear discriminants (LD). Each circle represents a cluster and each dot represents an individual. b Each single vertical line represents an individual and its proportional membership probability among the K clusters
Additionally, the FST pairwise comparisons were congruent with the previous results, showing no genetic differentiation between sympatric morpho-species (FST = 0.0003, P > 0.05). In contrast, allopatric populations showed highly significant FST values (P < 0.005). The differentiation levels between allopatric populations were higher than those observed between sympatric morpho-species from Lake Catemaco; thus, we obtain an FST = 0.202 for the comparison between A. aeneus from Lake Catemaco vs. A. aeneus from the Maquinas River. For the comparison of A. caballeroi from Lake Catemaco vs. A. aeneus from the Maquinas River, we obtained an FST = 0.200. Similarly, in our AMOVA, we found significant differentiation between geographic groups. Therefore, the only significant differences were between the Lake Catemaco vs. Maquinas River populations, also evidenced by the significant differences for the populations among groups; that is, there were significant differences between A. aeneus from Lake Catemaco and Maquinas River. In contrast, we did not find differences in the comparisons among sympatric populations, such as A. aeneus vs. A. caballeroi from Lake Catemaco (ESM Table S5).
Migration patterns between the two morpho-species. Migration rates and effective population size were estimated between sympatric A. aeneus and A. caballeroi. Both analyses, ML and BI, were concordant in the best model which corresponded to the third model (Ln ML = -266380.39 and Ln BF = -12879.64 respectively; Table 2). It is interesting to note that the migration rates between A. aeneus and A. caballeroi were asymmetric (Fig. 4), with the best model being the one that supports a migration from A. caballeroi to A. aeneus.
Migration rates, scaled by mutation rate between Astyanax aeneus and A. caballeroi. The circles are proportionally scaled to the theta values. The arrow's width is proportional to the M values. The A. aeneus mitochondrial lineage 1 presents a larger population size and the largest migration rates compared to the rest of the groups. Astyanax caballeroi species mitochondrial lineage 2 (A. caballeroi L2) shows the smallest population size and the lowest migration rates among the rest of the compared groups. The individuals were assigned to their mitochondrial lineage according to Ornelas-García et al. (2014)
Discussion
In the present study, we evaluated the degree of genetic differentiation between two morpho-species, Astyanax aeneus and A. caballeroi, currently recognized as valid and coexisting in Lake Catemaco, Mexico. The conspicuous phenotypic divergence between these morpho-species led to their original classification as different genera: Astyanax aeneus vs. Bramocharax caballeroi (Contreras-Balderas and Rivera-Teillery 1983). In this sense, we corroborated an association between their morphological and trophic divergence (Ornelas-García et al. 2014, 2018), suggesting an adaptive divergence process, in a context of ecological segregation. We found that A. caballeroi is a specialist species consuming more insects and with higher δ15N values, in contrast to the sympatric A. aeneus, which presents greater consumption of vegetal material (Ornelas-García et al. 2018).
Contrary to our hypothesis, the two species, A. aeneus and A. caballeroi, coexist in the presence of gene flow, evidenced by the admixture and high migration rates between them. At the same time, we found clear genetic differences with an allopatric population (i.e., Maquinas River vs. Lake Catemaco), supporting the notion that the marker system employed (i.e., microsatellites) has the resolution to capture genetic differences between populations within the same species. Despite this, we cannot rule out limitations in the microsatellite markers in discriminating incipient ecological divergence (see Kautt et al. 2016a, b). Thus, further studies, including genomic data, could help elucidate this effect (i.e., RAD-seq data).
On the other hand, we observed higher genetic diversity values in the Lake Catemaco system considering both morpho-species, in comparison to the allopatric Maquinas River population (Table 1). However, the diversity values recovered (i.e., Ho and He) were like those reported in other characid species (Beheregaray et al. 2005) and in its sister species A. mexicanus (Bradic et al. 2012).
We recovered a very low FST-value between the two sympatric species, A. aeneus vs. A. caballeroi (FST = 0.0003, P > 0.05). In a previous study, where pairs of lacustrine morphs of the genus Astyanax in Central America were compared, we obtained similar results (Garita-Alvarado et al. 2021). That is, in the comparisons between lacustrine sympatric ecotypes in Central America, we observed low, and for most of the comparisons, non-significant FST-values, except in the Río Sarapiquí system (FST = 0.022, P ≤ 0.05). Based on these results, we cannot rule out an incipient speciation process, or that our study system corresponds to a polymorphic species. Other systems, such as the Midas cichlid in Central America (i.e., Amphilophus tolteca), showed a very similar pattern. Based on 13 microsatellites, FST-values were very low between limnetic and benthic morphs (FST = 0.005 and P > 0.05) in Lake Asososca Managua (Kautt et al. 2016b). When demographic analyses based on RAD-seq data were integrated it was shown that this lack of differentiation was the result of a very recent divergence between these two sympatric morphs, in just a few hundred generations (Kautt et al. 2016b).
Phenotypic divergence with gene flow, on the way to speciation? In our study system, we did not find genetic differentiation between the two sympatric morpho-species. In contrast, we observed admixture between A. aeneus and A. caballeroi, corresponding to the same genetic cluster (Figs. 2, 3). Under an adaptive divergence scenario, the lack of genetic differentiation between sympatric divergent phenotypes could be due to either an incipient divergence process or to a stable polymorphism (e.g., Nosil et al. 2009; Magalhaes et al. 2015; Kautt et al. 2016a, b; Arbuthnott 2017). In this regard, the completion of the speciation process depends on multiple factors. For example, divergent selection depends on the strength of the selective forces on a trait (i.e., strong vs. weak) (Nosil et al. 2009; Arbuthnott 2017). Thus, loci under strong divergent selection could be expected to present higher levels of differentiation than neutral regions, which would show weak or absent differentiation (Barrett et al. 2008; Marchinko 2009; Nosil et al. 2009). Another factor is the extent of divergence along the genome, i.e., it could take place on a single locus vs. several loci (Jones et al. 2012; Nosil 2012; Kautt et al. 2020). Under a polygenic scenario (i.e., many loci), ecological speciation with gene flow could be the result of the genetic coupling between assortative mating and traits under divergent selection (Bay et al. 2017; Kautt et al. 2020). An alternative to the previous scenario is the single locus polymorphism, which does not always result in genome-wide divergence or ultimately in speciation. An example of this is found in the gold/dark morphs and lip-associated ecotypes in Midas Cichlids of the Great lakes in Central America (Kautt et al. 2020). In this previous study, the authors suggest that divergent selection affecting a large number of loci across the genome could result in post-zygotic isolation and promote irreversible barriers to gene flow under sympatric conditions. A similar pattern of polygenic barriers was suggested in the Heliconius butterfly species, in which under a gene flow scenario, the simultaneous selection across the genome could promote speciation (Feder and Nosil 2010; Martin et al. 2019). Therefore, using a small representation of markers may fail to provide enough information to characterize the genomic regions under divergence (Coyne and Orr 1998). Our study system opens the possibility to explore the genetic architecture of this phenotypic divergence under the previously described scenarios.
On the other hand, under an ecological divergence framework, reproductive isolation could be time dependent (Nosil 2012). Thus, recently diverged populations would show weaker signs of reproductive isolation (hence, genetic differentiation) than older systems. In this regard, our study system corresponds to one of the most recent pairs of divergent lacustrine morphotypes across the Mesoamerican region (Powers et al. 2020), whose lower phenotypic divergence is suggested to be related to the recent divergence time (Rosen 1972; Contreras-Balderas and Rivera-Teillery 1983; Ornelas-García et al. 2008; Powers et al. 2020. Thus, we could expect that the divergence between the morpho-species is indeed very recent, and therefore, no prezygotic barriers have evolved to prevent gene flow.
Finally, the coexistence of divergent phenotypes with gene flow has been shown in its sister group, A. mexicanus, although this gene flow is the result of a secondary contact process, it is interesting to note that despite this, the environment could be favoring certain phenotypic variants as shown in the study of Moran et al. (2022), observing a selection toward genes that could be relevant in the adaptation to caves. Our model system is very interesting to address in greater depth what could be these selective pressures that allow the presence of two divergent phenotypes in lacustrine systems, in the presence of gene flow.
Stable polymorphism as an alternative scenario. The two sympatric morpho-species could represent a case of stable polymorphism, where two divergent morphs develop out of a single gene pool, whereby the developmental trajectory to form one or both morphs would be determined during ontogeny (Garita-Alvarado and Ornelas-García 2021). Previous studies in fishes have provided some striking examples of stable polymorphic species. The Cuatro Cienegas Cichlid, Herichthys minckleyi, is a polymorphic species in body shape and trophic apparatus. It shows two pharyngeal jaw phenotypes: a molariform shape with a gastropod diet, and a papilliform morph, whose diet is composed of zooplankton, plants, and detritus (Hulsey et al. 2005; Magalhaes et al. 2015). In this sense, environmental clues such as frequency- or density-dependent selection could be the maintenance mechanism of this trophic polymorphism (Swanson et al. 2003). Another striking example of polymorphism in a fish species is the mouth asymmetry in the scale-eating cichlid Perissodus microlepis, from Lake Tanganyika, in which the direction of mouth opening (left-handed or right-handed) is determined by frequency-dependent selection imposed by the prey’s awareness (Hori 1993; Raffini et al. 2017). Notably, polymorphism is exhibited in several groups of insects (Nosil 2012). This mechanism explains how different morphs have arisen in some species of social insects (Ross and Keller 1995), usually coupled with monopolization of reproduction in only one of the morphs.
In our parallel evolved lacustrine morphotypes across Mesoamerica within the Astyanax genus, we assessed the genetic differentiation between another pair of sympatric divergent morphs in the Central American great lakes (i.e., Lake Managua and Lake Nicaragua). Despite their great ecological and morphological divergence (Powers et al. 2020), we found a marginal genetic differentiation and failed to recover discrete genotypic clusters based on microsatellite data (Garita-Alvarado et al. 2021). This suggests that stable polymorphism may be a plausible explanation for this ecomorphological divergence. However, additional analysis including genomic data is needed to understand the genetic architecture, as well as the selective mechanisms affecting the prevalence of these divergent phenotypes. Further crossbreeding of the morphotypes and/or performing common garden experiments could provide important information regarding the genetic basis of these phenotypes.
Taxonomic implications. In the present study, we observed the presence of gene flow between two sympatric morpho-species, A. aeneus and A. caballeroi, in Catemaco Lake, despite the marked phenotypic and ecological divergence between them (see Ornelas-García et al. 2017, 2018). Thus, considering both the biological species concept (Mayr 1963) and the genotypic grouped species concept (Mallet 1995), our two lake morpho-species could not be recognized as distinct, suggesting that our model system is at an early stage in the speciation process, or that it represents a polymorphic species. Elucidating the genetic basis underlying speciation is a long-standing goal of evolutionary biology, particularly in terms of delimiting a continuous process in a restricted temporal window (Simpson 1951; De Queiroz 2007). Therefore, the inclusion of genomic data could help to elucidate the genetic architecture of this phenotypic divergence and to better understand the mechanisms that allow its maintenance in the presence of gene flow.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Arbuthnott D (2017) The ecology of non-ecological diversification: how non-sexual selection affects within-environment diversification via sexual conflict. Evol Ecol Res 18:503–513
Barluenga M, Stölting KN, Salzburger W, Muschick M, Meyer A (2006) Sympatric speciation in Nicaraguan crater lake cichlid fish. Nature 439:719–723
Barrett RD, Rogers SM, Schluter D (2008) Natural selection on a major armor gene in threespine stickleback. Science 322:255–257
Bay RA, Arnegard ME, Conte GL, Best J, Bedford NL, McCann SR, Dubin ME, Chan YF, Jones FC, Kingsley DM (2017) Genetic coupling of female mate choice with polygenic ecological divergence facilitates stickleback speciation. Curr Biol 27:3344–3349
Beerli P, Felsenstein J (1999) Maximum-likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics 152:763–773
Beerli P, Felsenstein J (2001) Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. PNAS 98:4563–4568
Beerli P, Palczewski M (2010) Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics 185:313–326
Beheregaray L, Chae J, Chao N, Caccone A (2005) Characterization of microsatellite loci for the Amazonian rummy‐nose tetra, Hemigrammus bleheri (Teleostei, Characidae). Mol Ecol Notes 5:536–537
Belkhir K (2004) 1996–2004 GENETIX 4.05.2, logiciel sous Windows TM pour la genetique des populations. https://mybiosoftware.com/genetix-4-05-set-of-population-genetics-programs.html
Bolnick DI (2011) Sympatric speciation in threepine stickleback: why not? Int J Ecol 2011:942847
Bolnick DI, Snowberg LK, Patenia C, Stutz WE, Ingram T, Lau OL (2009) Phenotype-dependent native habitat preference facilitates divergence between Parapatric Lake and stream stickleback. Evolution 63:2004–2016
Bradic M, Beerli P, García-de Leon FJ, Esquivel-Bobadilla S, Borowsky RL (2012) Gene flow and population structure in the Mexican blind cavefish complex (Astyanax mexicanus). BMC Evol Biol 12:9
Contreras-Balderas S, Rivera-Teillery R (1983) Bramocharax (Catemaco) caballeroi subgen. et. sp. nv. del Lago de Catemaco, Veracruz, México (Pisces: Characidae). Inst Inv Cient UANL Mexico 2:7–29
Coyne JA, Orr AH (1998) The evolutionary genetics of speciation. Philos Trans R Soc Lond B Biol Sci 353:287–305
Criscuolo NG, Angelini C (2020) StructuRly: a novel shiny app to produce comprehensive, detailed and interactive plots for population genetic analysis. PloS One 15(2):e0229330
De Queiroz K (2007) Species concepts and species delimitation. Syst Biol 56:879–886
Dieckmann U, Doebeli M, Metz JA, Tautz D (2004) Adaptive speciation. Cambridge University Press, Cambridge
Doebeli M, Dieckmann U (2003) Speciation along environmental gradients. Nature 421:259–264
Elipot Y, Hinaux H, Callebert J, Launay JM, Blin M, Rétaux S (2014) A mutation in the enzyme monoamine oxidase explains part of the Astyanax cavefish behavioural syndrome. Nat Commun 5:3647
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Lischer HE (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Feder JL, Nosil P (2010) The efficacy of divergence hitchhiking in generating genomic islands during ecological speciation. Evolution 64:1729–1747
Garita-Alvarado CA, Barluenga M, Ornelas-García CP (2018) Parallel evolution of morphs of Astyanax species (Teleostei: Characidae) in México and Central America. Biol J Linn Soc Lond 124:706–717
Garita-Alvarado CA, Garduño-Sánchez MA, Barluenga M, Ornelas-García CP (2021) Genetic and ecomorphological divergence between sympatric Astyanax morphs from Central America. J Evol Biol 34:1752–1766
Garita-Alvarado CA, Ornelas-García CP (2021) Parallel evolution of allometric trajectories of trophic morphology between sympatric morphs of Mesoamerican Astyanax (Characidae). Appl Sci 11:8020
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices, version 2.9.3. http://www2.unil.ch/popgen/softwares/fstat.htm
Gross JB, Borowsky R, Tabin CJ (2009) A novel role for Mc1r in the parallel evolution of depigmentation in independent populations of the cavefish Astyanax mexicanus. PLoS Genet 5:e1000326
Guinand B (1996) Use of a multivariate model using allele frequency distributions to analyse patterns of genetic differentiation among populations. Biol J Linn Soc Lond 58:173–195
Guo SW, Thompson EA (1992) Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 48:361–372
Hatfield T, Schluter D (1999) Ecological speciation in sticklebacks: environment‐dependent hybrid fitness. Evolution 53:866–873
Hori M (1993) Frequency-dependent natural selection in the handedness of scale-eating cichlid fish. Science 260:216–219
Hulsey CD, Hendrickson DA, de León FJG (2005) Trophic morphology, feeding performance and prey use in the polymorphic fish Herichthys minckleyi. Evol Ecol Res 7:303–324
Jeffery WR (2009) Regressive evolution in Astyanax cavefish. Annu Rev Genet 43:25–47
Jombart T, Ahmed I, Jombart T, Ahmed I (2011) Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27:3070–3071
Jombart T, Collins C (2015) A tutorial for discriminant analysis of principal components (DAPC) using adegenet 2.0.0. MRC Centre for Outbreak Analysis and Modelling, Imperial College London, London
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94
Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MC, White S (2012) The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484:55–61
Kass RE, Raftery AE (1995) Bayes factors. J Am Stat Assoc 90:773–795
Kautt AF, Kratochwil CF, Nater A, Machado-Schiaffino G, Olave M, Henning F, Torres-Dowdall J, Häer A, Hulsey CD, Franchini P, Pippel M, Myers EW, Meyer A (2020) Contrasting signatures of genomic divergence during sympatric speciation. Nature 588:106–111
Kautt AF, Machado-Schiaffino G, Meyer A (2016a) Multispecies outcomes of sympatric speciation after admixture with the source population in two radiations of Nicaraguan crater lake cichlids. PLoS Genet 12(6):e1006157
Kautt AF, Machado-Schiaffino G, Torres-Dowdall J, Meyer A (2016b) Incipient sympatric speciation in Midas cichlid fish from the youngest and one of the smallest crater lakes in Nicaragua due to differential use of the benthic and limnetic habitats? Ecol Evol 6:5342–5357
Magalhaes IS, Ornelas-García CP, Leal-Cardin M, Ramírez T, Barluenga M (2015) Untangling the evolutionary history of a highly polymorphic species: introgressive hybridization and high genetic structure in the desert cichlid fish Herichtys minckleyi. Mol Ecol 24:4505–4520
Mallet J (1995) A species definition for the modern synthesis. Trends Ecol Evol 10:294–299
Marchinko KB (2009) Predation's role in repeated phenotypic and genetic divergence of armor in threespine stickleback. Evolution 63:127–138
Martin SH, Davey JW, Salazar C, Jiggins CD (2019) Recombination rate variation shapes barriers to introgression across butterfly genomes. PLoS Biol 17:e2006288
Mayr E (1963) Animal species and evolution. Animal species and evolution. Harvard University Press, Cambridge
McGaugh SE, Gross JB, Aken B, Blin M, Borowsky R, Chalopin D, Hinaux H, Jeffery WR, Keene A, Ma L, Minx P, Murphy D, Kelly E, O’Quin KE, Rétaux S, Ronher N, Sarle SM, Stahl BA, Tabin C, Volff JN, Yoshizawa M, Warren WC (2014) The cavefish genome reveals candidate genes for eye loss. Nat Commun 5:5307
McGaugh SE, Kowalko JE, Duboué E, Lewis P, Franz-Odendaal TA, Rohner N, Gross JB, Keene AC (2020) Dark world rises: The emergence of cavefish as a model for the study of evolution, development, behavior, and disease. J Exp Zool B Mol Dev Evol 334:397–404
Moran RL, Jaggard JB, Roback EY, Kenzior A, Rohner N, Kowalko JE, Ornelas-García CP, McGaugh S, Keene AC (2022) Hybridization underlies localized trait evolution in cavefish. iScience 25:103778
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Nei M, Chesser RK (1983) Estimation of fixation indices and gene diversities. Ann Hum Genet 47:253–259
Nosil P (2012) Ecological speciation. Oxford University Press, Oxford
Nosil P, Harmon LJ, Seehausen O (2009) Ecological explanations for (incomplete) speciation. Trends Ecol Evol 24:145–156
Ornelas-García CP, Bastir M, Doadrio I (2014) Morphometric variation between two morphotypes within the Astyanax Baird and Girard, 1854 (Actinopterygii: Characidae) genus, From a Mexican tropical lake. J Morphol 275:721–731
Ornelas-García CP, Bautista A, Herder F, Doadrio I (2017) Functional modularity in lake-dwelling characin fishes of Mexico. PeerJ 5:e3851
Ornelas-García CP, Córdova-Tapia F, Zambrano L, Bermúdez-González MP, Mercado-Silva N, Mendoza-Garfias B, Bautista A (2018) Trophic specialization and morphological divergence between two sympatric species in Lake Catemaco, Mexico. Ecol Evol 8:4867–4875
Ornelas-García CP, Domínguez-Domínguez O, Doadrio I (2008) Evolutionary history of the fish genus Astyanax Baird & Girard (1854) (Actinopterygii, Characidae) in Mesoamerica reveals multiple morphological homoplasies. BMC Evol Biol 8:340
Powers AK, Garita-Alvarado CA, Rodiles-Hernández R, Berning DJ, Gross JB, Ornelas-García CP (2020) A geographical cline in craniofacial morphology across populations of Mesoamerican lake‐dwelling fishes. J Exp Zool A Ecol Integr Physiol 333:171–180
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Protas M, Conrad M, Gross JB, Tabin C, Borowsky R (2007) Regressive evolution in the Mexican cave tetra, Astyanax mexicanus. Curr Biol 17:452–454
Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, Jeffery WR, Zon IZ, Borowsky R, Tabin C (2006) Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet 38:107–111
Raffini F, Fruciano C, Franchini P, Meyer A (2017) Towards understanding the genetic basis of mouth asymmetry in the scale‐eating cichlid Perissodus microlepis. Mol Ecol 26:77–91
R Core Team (2022) R: a language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria. https://www.R-project.org
Raymond M (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43: 223–225
Rosen DE (1972) Origin of the characid fish genus Bramocharax and description of a second, more primitive, species in Guatemala. Am Mus Novit 2500:1–21
Ross KG, Keller L (1995) Ecology and evolution of social organization: insights from fire ants and other highly eusocial insects. Annu Rev Ecol Syst 26:631–656
RStudio Team (2022) RStudio: integrated development for R. RStudio, PBC, Boston. http://www.rstudio.com/
Schliewen U, Rassmann K, Markmann M, Markert J, Kocher T, Tautz D (2001) Genetic and ecological divergence of a monophyletic cichlid species pair under fully sympatric conditions in Lake Ejagham, Cameroon. Mol Ecol 10:1471–1488
Schluter D (1996) Ecological causes of adaptive radiation. Am Nat 148:S40–S64
Schmitter-Soto JJ (2017) A revision of Astyanax (Characiformes: Characidae) in Central and North America, with the description of nine new species. J Nat Hist 51:1331–1424
Simpson GG (1951) The species concept. Evolution 5:285–298
Sonnenberg R, Nolte AW, Tautz D (2007) An evaluation of LSU rDNA D1-D2 sequences for their use in species identification. Front Zool 4:6
Swanson BO, Gibb AC, Marks JC, Hendrickson DA (2003) Trophic polymorphism and behavioral differences decrease intraspecific competition in a cichlid, Herichthys minckleyi. Ecology 84:1441–1446
Thibert-Plante X, Hendry A (2009) Five questions on ecological speciation addressed with individual‐based simulations. J Evol Biol 22:109–123
Van Oosterhout C, Hutchinson WF, Wills DP, Shipley P (2004) MICRO‐CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
West-Eberhard MJ (2003) Developmental plasticity and evolution. Oxford University Press, Oxford
Acknowledgements
The authors thank: Carlos Pedraza for field and laboratory assistance and his helpful suggestions on an early version of the manuscript; Lourdes Alcaraz and Silvia Perea for their helpful suggestions on the early version of the article and laboratory assistance. We acknowledge funding sources from Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (IN212419) to CPOG and Max Planck Institute for Evolutionary Biology in Plön Germany.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human and animal rights
In our research, no laboratory experiments with animals were performed. The fishes collected during the fieldwork were immediately euthanized in cool water to minimize the suffering of the animals and to meet acceptable animal ethical standards. Fish were collected under permits SGPA/DGVS/05464/, SAGARPA (México).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Ornelas-García, C.P., Gonzalez, E.G., Tautz, D. et al. Lack of genetic differentiation between two sympatric lacustrine morpho-species within the Astyanax (Characidae: Teleostei) genus, Mexico. Ichthyol Res 71, 95–105 (2024). https://doi.org/10.1007/s10228-023-00917-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10228-023-00917-1