Abstract
Background
The European Society for Medical Oncology Precision Medicine Working Group (ESMO-PMWG) published recommendations regarding confirmatory germline testing for presumed germline pathogenic variants (PGPVs) in tumor-only comprehensive genomic profiling (CGP). However, the clinical validity of these recommendations has not been investigated in a real-world practice.
Methods
Medical records of 180 consecutive patients who obtained the results of a tumor-only CGP (FoundationOne® CDx, Foundation Medicine, Inc, Cambridge, MA, USA) between October 2018 and March 2020, were retrospectively reviewed. After excluding patients with no reported variants in 45 actionable genes (n = 6), or no archived germline DNA samples (n = 31), 143 patients were investigated. The PGPVs were selected from the CGP report and germline sequencing were performed using DNA samples archived in Clinical Bioresource Center in Kyoto University Hospital (Kyoto, Japan).
Results
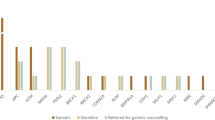
A total of 195 variants were classified as PGPV based on the conventional criteria. Germline sequencing disclosed that 12 variants (6.2%) were of germline origin. In contrast, after filtering these 195 variants through the ESMO-PMWG recommendation criteria for confirmatory germline testing, following seven PGPVs, BRCA2 (n = 2), BRIP1 (n = 1), BAP1 (n = 1), PMS2 (n = 1), MSH2 (n = 1), and SDHB (n = 1) remained and six variants (85.7%) were confirmed to be of germline origin.
Conclusion
Our current data suggested that the application of ESMO-PMWG criteria is helpful in selecting PGPVs with a high likelihood of germline origin in a tumor-only CGP in daily clinical practice.




Similar content being viewed by others
References
Haendel MA, Chute CG, Robinson PN (2018) Classification, ontology, and precision medicine. N Engl J Med 379:1452–1462
Li MM, Chao E, Esplin ED et al (2020) Points to consider for reporting of germline variation in patients undergoing tumor testing: a statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med 22:1142–1148
Mandelker D, Donoghue M, Talukdar S et al (2019) Germline-focussed analysis of tumour-only sequencing: recommendations from the ESMO Precision Medicine Working Group. Ann Oncol 30:1221–1231
Frampton GM, Fichtenholtz A, Otto GA et al (2013) Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 31:1023–1031
Kondo T, Matsubara J, Quy PN et al (2021) Comprehensive genomic profiling for patients with chemotherapy-naïve advanced cancer. Cancer Sci 112:296–304
Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405–424
Kalia SS, Adelman K, Bale SJ et al (2017) Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med 19:249–255
Pujol P, Vande Perre P, Faivre L et al (2018) Guidelines for reporting secondary findings of genome sequencing in cancer genes: the SFMPP recommendations. Eur J Hum Genet 26:1732–1742
Cingolani P, Platts A, le Wang L et al (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92
Coombs CC, Zehir A, Devlin SM et al (2017) Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell 21:374-382.e374
Landrum MJ, Lee JM, Benson M et al (2018) ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 46:D1062-d1067
Li Q, Wang K (2017) InterVar: clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am J Hum Genet 100:267–280
Hayashi H, Tanishima S, Fujii K et al (2020) Clinical impact of a cancer genomic profiling test using an in-house comprehensive targeted sequencing system. Cancer Sci 111:3926–3937
Vlessis K, Purington N, Chun N et al (2019) Germline testing for patients with BRCA1/2 mutations on somatic tumor testing. JNCI Cancer Spect. https://doi.org/10.1093/jncics/pkz095
Friedman JM (1993) Neurofibromatosis 1. In: Adam MP, Ardinger HH, Pagon RA et al (eds) GeneReviews(®). University of Washington, Seattle
Giusti F, Marini F, Brandi ML (1993) Multiple Endocrine Neoplasia Type 1. In: Adam MP, Ardinger HH, Pagon RA et al (eds) GeneReviews(®). University of Washington, Seattle
Jaiswal S, Libby P (2020) Clonal haematopoiesis: connecting ageing and inflammation in cardiovascular disease. Nat Rev Cardiol 17:137–144
Silver AJ, Bick AG, Savona MR (2021) Germline risk of clonal haematopoiesis. Nat Rev Genet. https://doi.org/10.1038/s41576-021-00356-6
Ptashkin RN, Mandelker DL, Coombs CC et al (2018) Prevalence of clonal hematopoiesis mutations in tumor-only clinical genomic profiling of solid tumors. JAMA Oncol 4:1589–1593
Lincoln SE, Nussbaum RL, Kurian AW et al (2020) Yield and utility of germline testing following tumor sequencing in patients with cancer. JAMA Netw Open 3:e2019452–e2019452
Acknowledgements
The authors would like to thank the patients for their kind cooperation, as well as Mari Funakoshi, Hitomi Sakamoto, Kanami Ashida, Junko Suga, Momoko Sato, Kumi Mukai, and all staff members of Clinical Bioresource Center in Kyoto University Hospital for their excellent technical and secretarial assistance.
Funding
This study was supported by the Japan Agency for Medical Research and Development, AMED, under Grant number 17kk0305006h0001. This research received funding from Chugai Pharmaceutical Co., Ltd., Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Masashi Kanai received lecture fees, honoraria, and other fees from Chugai Pharmaceutical Co., Ltd. Atsushi Yamada belong to an endowed chair sponsored partly by Chugai Pharmaceutical Co., Ltd. Manabu Muto received research funding, lecture fees, honoraria, and other fees from Chugai Pharmaceutical Co., Ltd. All remaining authors have declared no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
About this article
Cite this article
Kondo, T., Yamamoto, Y., Fukuyama, K. et al. Germline sequencing for presumed germline pathogenic variants via tumor-only comprehensive genomic profiling. Int J Clin Oncol 27, 1256–1263 (2022). https://doi.org/10.1007/s10147-022-02176-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10147-022-02176-y