Abstract
Cuproptosis is a newly discovered form of cell death. It is regulated by a string of genes. The genes are identified to influence the tumor progression, but in glioma, the cuproptosis-related genes are little studied. The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) were used to screen for SLC31A1 gene expression in glioma and healthy tissue samples. The results were validated using the Gene Expression Omnibus (GEO) and quantitative real-time polymerase chain reaction (qPCR). The Human Protein Atlas (HPA) and the National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC) were used to validate our results at the protein level. Multivariable analysis and Kaplan–Meier survival curves were used to examine the relationship among SLC31A1 gene expression, clinical parameters, and survival rates. The online Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) was used to find the genes and proteins that correlate to SLC31A1. The immune infiltration analysis was performed using the Tumor Immune Estimation Resource (TIMER) databases. Small interfering RNA was used to knock down the SLC31A1 expression, and the cell proliferation, apoptosis, and migration were analyzed using cell counting kit-8, flow cytometry, and transwell. The glioma patients have higher SLC31A1 expression levels, which increase as the World Health Organization (WHO) grade escalates. The survival analysis illustrates that the SLC31A1 gene expression negatively correlates with overall survival (OS), progression-free survival (PFS), and disease-specific survival (DSS). The immune infiltration analysis shows the SLC31A1 gene positively correlates with T helper 2 (Th2) cells, macrophages, and M2-type macrophages and negatively correlates with plasmacytoid dendritic cells (pDCs), natural killer (NK) CD56bright cells, and CD8 T cells. The in vitro KD experiment shows the SLC31A1 knockdown depressed the glioma cell proliferation and migration and promoted the apoptosis rate. The SLC31A1 gene expression can shorten the survival time of glioma patients. In vitro study shows that SLC31A1 can promote cell proliferation, and migration, and depress the cell apoptosis of glioma cells. It also can promote the formation of a tumor-suppressive microenvironment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Cuproptosis is a recently discovered form of programmed cell death regulated by a set of genes known as cuproptosis-correlated genes (CRGs). While several studies have explored the role of CRGs in various cancers, the extent of research on the SLC31A1 gene is limited. Glioma, a type of brain tumor, is known to be associated with dysregulated cell death pathways, making it a prime candidate for investigation of the role of SLC31A1.To this end, we analyzed data from online databases to investigate the relationship between SLC31A1 gene expression and clinical outcomes and the tumor microenvironment in glioma patients. Our results revealed that high expression of the SLC31A1 gene is associated with unfavorable outcomes and an immunosuppressive tumor microenvironment. These findings suggest that SLC31A1 could be a valuable target for further investigation in the development of novel therapies for glioma.
Introduction
Gliomas are the most common primary tumors in the brain, accounting for 81% of central nervous system (CNS) malignancies (Ostrom, et al. 2018). The current standard therapy interventions, including surgery, radiotherapy, and systemic chemotherapy, cannot convert the most treatment resistance of glioma (Jiang et al. 2021; Armstrong et al. 2020). One of the many reasons is the glioma immunosuppressive tumor microenvironment (TME) (Quail and Joyce 2017). As previously reported, the TME of glioma is immunosuppressive and is known as one of the cold immune tumors (D'Alessio, et al. 2019; Xiong et al. 2022). More key molecules need to be identified to understand the TME of glioma and improve the outcomes of glioma patients.
Copper is an essential element in cells (Lewinska-Preis et al. 2011). There is a balance in copper metabolism, and if this balance is broken, it may cause cell death, called cuproptosis. Furthermore, the scientists revealed that the cuproptosis is regulated by a series of genes, including FDX1, LIPT1, LIAS, DLD, DBT, DLAT, GCSH, DLST, ATP7A, ATP7B SLC31A1 (solute carrier family 31 member 1), PDHA1, and PDHB (Tsvetkov et al. 2022). Most genes, such as FDX1, have been studied in different cancers to different degrees (Wang et al. 2022a; Zhang et al. 2022a; Zhang et al. 2022b; Song et al. 2022). Besides, we noticed that SLC31A1 had been studied in different cancers and strongly correlated with poor prognosis (Li et al. 2022a; Kong et al. 2023). However, in glioma, the SLC31A1 has not been studied in detail.
In this study, we used data from public datasets, including the Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), the Human Protein Atlas (HPA), the Chinese Glioma Genome Atlas (CGGA), and the Clinical Proteomic Tumor Analysis Consortium (CPTAC) to investigate the association between SLC31A1 gene expression and clinical outcomes in glioma patients. Then, we tested the SLC31A1 gene expression difference between glioma cell lines U87, LN229, U251, U343, and the average human fetal glial cell line SVGp12 using a Real-time Quantitative PCR Detecting System. We then used data from the Tumor Immune Estimation Resource (TIMER) and the Gene Expression Profiling Interactive Analysis (GEPIA) databases to analyze the relationship between SLC31A1 gene expression and immune cell infiltration and the associated gene marker sets. Furthermore, we analyzed the SLC31A1-interacting protein network using the STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) platform. A high SLC31A1 gene level is related to infiltrating immune cell changes in glioma tissues, indicating a dismal prognosis. Thus, it is plausible that the SLC31A1 gene defect may debilitate antitumor immune effects for glioma. SLC31A1-related targeting may be a viable treatment approach in glioma in combination with immunotherapy.
Results
SLC31A1 gene expression was increased in glioma as opposed to normal samples
To determine whether SLC31A1 gene expression is higher in glioma patients, we analyzed data from different datasets and levels; at first we analyzed the SLC31A1 gene expression in glioma samples using the data from the TCGA dataset and compared it to the normal tissue samples from the GTEx dataset (Fig. 1A) (N = 1157, T = 689). The GEO datasets were used to test the accuracy of the tendency (Fig. 1B) (N = 23, T = 153). As demonstrated, the SLC31A1 gene expression at the mRNA level is upmodulated in glioma patients (p < 0.001) (Fig. 1A and B). It has been shown that the SLC31A1 gene may play a key role in glioma. Then, we further tested the SLC31A1 protein expression level by using data from the CPTAC database (N = 10, T = 100), and we found that the SLC31A1 protein is upmodulated in glioma samples (p = 0.0205) (Fig. 1C). These results support that the SLC31A1 is a glioma-promotive gene, as the previous study has partially put it (Li, et al. 2023). Cell lines were also used to confirm the difference in expression between glioma and normal control cell lines. We found that the SLC31A1 gene expression is significantly upregulated (p < 0.0001) in glioma cell lines (Fig. 1D). Additionally, to further determine the expression level of SLC31A1 at the protein level, we used the histochemical data from HPA. As is shown in Fig. 1E and 1F, the positive rates of SLC31A1 are higher in HGG (high-grade glioma) patients than in LGG (low-grade glioma) patients. All of the results indicate that the SLC31A1 gene is associated with glioma (Table 1).
Association between SLC31A1 expression and clinical parameters
Based on the previous data, it has been found that SLC31A1 is upmodulated in glioma patients. Therefore, we investigated the difference in SLC31A1 gene expression in patients with different clinical parameters, based on the expression differences of SLC31A1, we separated the patients into low- and high-expression groups. The clinicopathological characteristics in patients with glioma are shown in Table 2. We used the Kruskal–Wallis and Wilcoxon signed-rank tests to determine the relationship between SLC31A1 expression and clinical parameters. We found that higher WHO grade (G2 = 224, G3 = 243, G3 = 168), higher age (≤ 60 = 553, > 60 = 143), and progressive disease (PD) (PD = 112, SD = 147, PR = 64, CR = 139) were associated with high-level SLC31A1 expression (Fig. 2A, B, and C). The SLC31A expression level escalates as the WHO grade increases. The results indicate the SLC31A1 gene is correlated with bad outcomes in glioma patients. Also, the SLC31A1 expression level is higher in patients with IDH WT status (WT = 246, Mut = 440) and 1p/19q non-codeletion (codel = 171, non-codel = 518), which were proven to have a strong correlation with unfavorable prognostic outcomes in glioma patients (p < 0.01) (Fig. 2D and E). The data from CGGA confirmed the same tendency (Figure S1). Nevertheless, there is no significant SLC31A1 difference between male (398) and female (298) patients (Fig. 2F). Overall, our analysis demonstrated that the SLC31A1 gene expression positively correlates with WHO grades, patients’ age, and bad primary therapy outcomes, which are indicators of poor outcomes. These results confirm SLC31A1 gene has a tremendous prognostic value in glioma patients.
Prognostic relevance of SLC31A1 expression in glioma
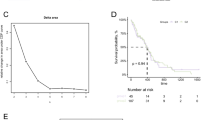
The former results show that the SLC31A1 gene is associated with the prognostic parameters; we then further investigated the prognostic survival value of SLC31A1 in glioma patients. We ranked the TCGA glioma patient samples by the expression level of SLC31A1, and the glioma cohort was classified into low- and high-expression groups according to the expression levels of SLC31A1. Then, we compared the prognosis difference between the two groups. As the pictures show, the survival curves of the low SLC31A1 group are above the high SLC31A1 group in OS, DSS, and PFS, which means the low SLC31A1 group survives longer than the high SLC31A1 group (Fig. 3A, B, C) (low = 348, high = 347), the data from CGGA shows the same tendency (Figure S2). The results mean that the high-level expression of SLC31A1 was associated with unfavorable OS (hazard ratio (HR) = 3.19(2.45–4.15), p < 0.001, Fig. 3A), DSS (HR = 3.45 (2.60–4.58), p < 0.001, Fig. 3B), and PFS (HR = 2.44 (1.95–3.04), p < 0.001, Fig. 3C). Then, a receiver operating characteristic (ROC) curve was constructed to examine the diagnostic significance of SLC31A1 expression by comparing SLC31A1 expression in normal tissue specimens (data obtained from GTEx) and glioma tissues (from the TCGA database). As is demonstrated that the area under the curve (AUC) value for SLC31A1 levels was 0.968 (confidence interval = 0.960–0.977), which means strong potential for diagnostic use (Fig. 3D) (N = 1157, T = 689). Following that, a clinical prognostic risk score for glioma was created using age, WHO grade, IDH status, 1p/19q codeletion, primary therapy outcome, and SLC31A1 expression. We found that the SLC31A1 has great prognostic predicted efficiency in glioma patients (Fig. 3E). Similarly, we also draw the same conclusion in univariate and multivariate analyses (using the Cox regression model) (Figure S3). In this part, we found that the SLC31A1 is an independent risk factor of glioma patients, which is useful for the outcome prediction of glioma. But how this gene acts is still unclear; we then further investigated the functions of SLC31A1.
The patients with higher SLC31A1 expression is more likely to have a worse prognosis. Patients with high SLC31A1 expression had unfavorable prognosis indicators than patients with low SLC31A1 expression, including shorter overall survival (OS) (A), progression-free interval (DSS) (B), and disease-specific survival (PFS) (C) (Log-rank p < 0.001). Receiver operating characteristic curve for SLC31A1 expression in normal samples (obtained using GTEx data) and adjoining glioma tissues and samples (D). A multivariate analysis nomogram based on clinical features associated with SLC31A1 expression (E)
The functions of SLC31A1
To better understand the functions of the SLC31A1 gene in glioma, we analyzed the genes related to the SLC31A1 by the STRING network. As is illustrated in Fig. 4A, the most top 10 genes related to SLC31A1 are ATOX1, ATP7B, ATP7A, COX17, CCS, SLC11A2, MTF1, SLC22A2, ZBED3, and CP. Annotations and their co-expression scores are displayed in Table 1. We then used the GO-KEGG enrichment analysis to analyze the functions of these genes. We found that the genes are mainly enriched to copper ion transportation pathways (Fig. 4B), showing that the SLC31A1 is a key gene for Cu2+ concentration regulation, which can be supported by a former study (ZHOU, B. and J. GITSCHIER 1997). To better understand the biological role of the SLC31A1 gene in glioma, we analyzed the whole-genome profile of glioma patients from TCGA. It was discovered that 285 genes are downregulated, and 1686 genes are upregulated, linked to the SLC31A1 gene expression. The gene expression heat map displayed the top 40 aberrant gene expression levels (logFC > 1.5 and padj < 0.05) (Fig. 4D). To further investigate the possible functions of SLC31A1 in glioma, the enrichment analysis was performed based on the SLC31A1 gene expression results. The BP primarily associated with the SLC31A1 gene was the regulation of response to food, antimicrobial humoral response, regulation of response to extracellular stimulus, regulation of response to nutrient levels, and defense response to the bacterium, among others (Fig. 4E). The results show a strong correlation to immune response, which is a strong influence factor of glioma progression; then, we further investigated the correlation between SLC31A1 and the TME of glioma.
The functions of SLC31A1. The SLC31A1-correlated genes from STRING database (A). The GO/KEGG analyses of the SLC31A1-correlated genes (B). The gene expression difference correlates to SLC31A1 in glioma (C). The top 40 genes correlate to SLC31A1 (D). The GO and KEGG analyses using the top correlating genes
The association between the SLC31A1 gene expression and immune cell infiltration
As previously described, the genes associated with SLC31A1 analyzed have shown a strong correlation with immunity (Li et al. 2022b). We then explored the relationship between SLC31A1 gene expression and 24 distinct immune subtypes in glioma using the online analysis tool of the TIMER2.0 database. We found that the SLC31A1 gene expression had a positive correlation with Th2 cells, macrophages, aDC cell infiltration, and a strong inverse correlation with pDC, NK CD56bright cells, and cytotoxic CD8 + T cell infiltration, among other cells (Fig. 5A, B–D). In the TME of glioma, the tumor-associated macrophages (TAMs) are critical; their volume may take up 30% of the glioma tissue (Hambardzumyan et al. 2016); the macrophages can be divided into M0, M1, and M2 types by the biomarkers and functions they expressed. Previous studies demonstrated that the M1-type macrophage is immune-promotive (Wang et al. 2022b) and the M2-type macrophage is immune-suppressive (Yunna et al. 2020). Th2 cells are also important tumor-promoting cells that can promote the M2 polarization of macrophage by secreting IL4 and IL13 cytokines (Gordon and Martinez 2010; Stark et al. 2019). As we investigated, the Th2 cell infiltration positively correlates to SLC31A1 gene expression. The M2-type macrophages are also upregulated as the SLC31A1 gene expression increases, indicating that the SLC31A1 may promote macrophage polarization by promoting the IL4 and IL13 secretion of Th2 cells. Further investigation illustrated that the SLC31A1 gene expression level differs in immune cell infiltration; the most correlated are neutrophils, eosinophils, T cells, and T helper cells, among others (Fig. 5B–D). To investigate the possible function of the SLC31A1 gene in influencing the infiltration status of different immune cells in glioma, we analyzed data from the TIMER and GEPIA databases to uncover the link between the SLC31A1 gene and different immune cell infiltration, including DCs, NK cells, M1/M2 macrophages, T cells (general), neutrophils, tumor-associated macrophages (TAMs), B cells, monocytes, and CD8 + T cells, in glioma. Besides, this study also evaluated different T cell subtypes, such as exhausted T cells, Tregs, Th1, Th2, Th9, Th17, Th22, and TFh. According to our results, the expression of most immune cell markers for various types of M1/M2 macrophages, TAMs, DCs, and T cells is linked to the SLC31A1 gene expression level in glioma. Among them, the Th2 and macrophage cells are the most relevant cells, which shows that the SLC31A1 gene may have a strong influence on the TME of glioma.
SLC31A1 knocks down depressed glioma proliferation and migration, promoted the glioma apoptosis
As SLC31A1 is crucial in glioma, we then tested the SLC31A1 gene function by in vitro experiments using LN229 and U251 cell lines. We knocked down (KD) the SLC31A1 in LN229 and U251 by small interferon RNA (siRNA), the Si504 shows strong knockdown efficiency than the other two siRNAs in both LN229 (95.7%) and U251 (95.3%) cell lines (Fig. 6A and Figure S4 A). Then, we analyzed the cell proliferation, apoptosis, and migration after knocking down the SLC31A1. Compared to the NC group, the si504 group shows a slower growth speed in both LN229 and U251 (Fig. 6B and Figure S4 B), and the flow cytometry assay shows the si SLC31A1 group has more apoptotic cell rates than the NC group (Fig. 6B and C, Figure S4 B & C). The transwell assay also shows the si504 group has less penetrated cells (Fig. 6E and F, Figure S4 E & F). The results show that the proliferation and migration were both depressed in LN229 and U251. KD SLC31A1 also promoted glioma cell apoptosis. This means the SLC31A1 may function by promoting the proliferation and migration of glioma cells. And the SLC31A1 gene might be important for the resistance of apoptosis in glioma. The experiments were repeated for 3 times and the data were processed by t test using Prism. A p value < 0.05 was considered statistically significant (*p < 0.05; **p < 0.01; ***p < 0.001; and ****p < 0.0001).
SLC31A1 knockdown depressed glioma viability and migration. We knocked down (KD) the SLC31A1 gene expression in glioma cell line LN229 by siRNA and the KD efficiency was detected by the qPCR method (A). The cell proliferation difference was detected by the CCK-8 Kit (B). The SLC31A1 KD promoted glioma cell apoptosis (C, D). The cell migration rate was analyzed by transwell (E, F). All the experiments have been repeated for three times
Discussion
Glioma is a refractory disease. Currently, our cognition of glioma is still limited. Scientists are trying to find new ways to understand and treat this deathful disease. Many factors are considered to be very important in glioma progression, such as gene mutation (Eckel-Passow et al. 2015), metabolism (Bi et al. 2020; Yu et al. 2023), tumor microenvironment (Friebel et al. 2020), and many kinds of cell death (Liu et al. 2022; Li et al. 2023; Abulaiti et al. 2023). Cuproptosis is a novel mode of cell death, especially in tumor cells; recent studies show that cuproptosis occurs in tumors and strongly impacts patients’ outcomes. Further research has revealed that cuproptosis can promote immune-suppressive microenvironment formation in different cancers (Du et al. 2022; Lv et al. 2022; Wang et al. 2022c). But only a few studies have examined its effects on glioma. The SLC31A1 encodes a membrane protein that can transport copper ions into the cells (ZHOU, B. and J. GITSCHIER 1997). It promotes copper ion metabolism in glioma cells; as the former research shows, the SLC31A1 gene is an essential gene for cuproptosis. We hypothesize that the SLC31A1 gene can promote the progression of glioma. As we analyzed, the high-SLC31A1 group glioma patients had worse outcomes than those with lower SLC31A1 gene expression levels. This finding might indicate that the glioma cell can resist cuproptosis by transporting the Cu2+ into the cells, which may be the molecular function of the SLC31A1 that contributes to glioma progression.
The previous study showed that the IDH mutation and 1p/19q codeletion significantly correlate with better outcomes in glioma patients (Sasmita et al. 2018; Jenkins et al. 2006). So we analyzed the relationship between SLC31A1 expression level and IDH mutation, 1p/19q codeletion. We found that the SLC31A1 expression level is lower in the IDH mutation group and 1p/19q codeletion group, which means that the SLC31A1 might influence the outcomes of glioma patients by influencing IDH mutation and 1p/19q codeletion.
In our research, we also explored the relationship between SLC31A1 gene expression and the outcomes of glioma patients. The high SLC31A1 expression level strongly correlates with bad outcomes in glioma patients. Following that, we further investigated the possible mechanism of how the SLC31A1 gene expression influences glioma progression. The in vitro knockdown experiment also shows that the SLC31A1 KD strongly impaired the proliferation and migration of glioma cells, which is the same as we analyzed by the in silico data. This means the SLC31A1 may promote glioma progression by enhancing the proliferation and migration of glioma cells. After that, we sorted the top 40 genes that mostly correlate with the SLC31A1 gene in glioma. Then, we did a gene enrichment analysis by using these genes. The result indicates that these genes have a strong correlation with immune response. That means the SLC31A1 gene might influence the glioma progression by reshaping the immune microenvironment. As former researches show (Quail and Joyce 2017; Yu et al. 2023; Klemm, et al. 2020), the TME of glioma plays a significant role in glioma progression. The former research has illustrated that the TME of glioma is suppressive, and the suppressive TME can promote the immune escape of glioma cells besides, as the brain is an immune-privileged organ. The TME of glioma is complicated, there are lots of investigations about it, and scientists have found many regulating factors of the glioma TME. As we investigated, the cuproptosis-correlated gene SLC31A1 is related to the immune response, but whether and how it acts in the TME of glioma is not clearly investigated. So, we then investigated the correlation between immune cell infiltration and SLC31A gene expression. We found that the SLC31A1 gene influences many kinds of immune cell infiltration. Mostly the tumor-associated macrophages (TAMs), T helper 2 (Th2) cells, plasmacytoid dendritic cells (pDCs), NK CD56bright cells, and CD8 T cells. Tumor-associated macrophage (TAM) refers to the macrophages recruited in the tumors; it also includes the three types of macrophages mentioned. Microglia and macrophages are the main component of glioma tumor–associated macrophages (TAMs) and have been categorized as M1 polarized (“classically activated”), M2 polarized (“alternatively activated”), and nonpolarized M0 macrophages (Ochocka et al. 2021). Among them, M0 macrophages are defined as undifferentiated macrophages with the potential to polarize into specific macrophage subtypes; M1 macrophages are classically activated, typically by IFN-γ or lipopolysaccharide (LPS), and produce proinflammatory cytokines, phagocytize microbes, and initiate an immune response (Mills, et al. 1950); M2 macrophages mainly secrete arginase-I, IL-10, and TGF-β and other anti-inflammatory cytokines, which have the function of reducing inflammation and contributing to tumor growth and immunosuppressive function (Hambardzumyan et al. 2016; Mosser and Edwards 2008; Roszer 2015). As we analyzed, in GBM, the SLC31A1 gene expression positively correlates with M2 macrophage infiltration but negatively correlates with M1 macrophage infiltration, which may cause immune-suppressive TME formation and promote the immune escape of the glioma cells. Th cells can also influence the TME of glioma; they are divided into different subtypes by the cytokines they secrete, mostly the Th1 and Th2 cells (Zhu and Zhu 2020). As previous research described, the Th2 bias, which means the Th2 ratio increasing, is important for glioma progress. The Th2 cell infiltration level increases as the WHO grades increase (Shimato 2012). The Th2 cells can secret IL4 and IL13 cytokines with strong M2 polarization functions of macrophages (Walker and McKenzie 2018). Another study found that Th2 bias in glioma is associated with increased expression of STAT6, a transcription factor that regulates Th2 differentiation (Shimato 2012). In short, in glioma, the M2-type macrophages are proven to have a strong immune suppressive function (Vidyarthi et al. 2019; Xu et al. 2021). As we investigated, the SLC31A1 gene expression positively correlates with Th2 cells, which indicated that the SLC31A1 has a strong impact on the TME of glioma. However, the study has limitations. Firstly, the function of the SLC31A1 gene in the glioma microenvironment is still not clear. Further investigations need to be done to understand the SLC31A1 gene. Secondly, how the SLC31A1 acts in glioma are still unclear, the analysis based on online data shows that the SLC31A1 influenced the progression of glioma, but whether and how it regulates the cuproptosis and other cell physiological activities in glioma is not validated, the further investigation is needed.
Conclusions
Our results indicate that the SLC31A1 gene expression can promote the formation of an immune-suppressive glioma microenvironment. The results also show that the SLC31A1 might be a critical molecular for the immune regulation of glioma. The medications targeting SLC31A1 may be helpful for the treatment of glioma. Fully understanding the SLC31A1 may be beneficial for clinical staging and personalized therapy.
Materials and methods
Cell lines
The human glioma cell lines U87, LN229, U251, and U343 and the normal human fetal glial cell line SVGp12 were purchased from ATCC, and all cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) with 10% FBS (PAN, Germany) in a humidified chamber at 5% CO2 and 37 ℃.
Data sources
The TCGA database
The TCGA (https://portal.gdc.cancer.gov) is a public data platform for cancer genome projects. It provides clinical and pathological data on 33 types of cancer that are easy to obtain. We searched the TCGA database for clinical data and high-throughput RNA sequencing (RNA-seq) information on patients with glioma. Moreover, we used fragments per kilobase per million fragments mapped (FPKM) included in HTSeq to determine SLC31A1 transcript expression levels. Furthermore, to further investigate the RNA-Seq gene expression level 3 HTSeqFPKM data of 703 patients with glioma, the clinical data were transformed into transcripts per million (TPM) reads. The database is public; therefore, the local ethics committee has no permission.
The CGGA database
The CGGA (https://www.cgga.org.cn/) is a public data platform that has collected more than 2000 Chinese glioma patients’ clinical and gene expression data. We used the data from CGGA to test the conclusions drawn from the TCGA database.
The GEO database
The GEO database (https://www.ncbi.nlm.nih.gov/geo/) is a public functional genomics data repository that supports MIAME-compliant data submissions. It is a comprehensive gene expression bank at the National Center for Biotechnology Information (NCBI). It provides gene expression data and clinical data of different patients, including glioma patients and healthy individuals. We used the data from the GEO database to examine the differences in SLC31A1 expression levels between glioma and normal tissues.
The CPTAC database
The CPTAC database (https:// proteomics.cancer.gov/programs/cptac/) is proteogenomic, which can provide proteome expression data of many kinds of tumors and normal tissues, including glioma and normal brain tissue. We downloaded the SLC31A1 protein expression data and analyzed the expression difference between normal brain tissue and glioma.
The HPA database
The HPA database (https://www.proteinatlas.org/) is a giant online database that contains extensive transcriptome and proteome information of different human specimens, including tissue, cell, and pathology samples. The HPA database can provide data on the cell-specific positions of the 44 normal tissues and the twenty most susceptible cancers. Moreover, the database provides information on protein immunohistochemistry in tumors and standard human tissue samples.
Statistical analysis of clinical prognosis, model development, and assessment
We analyzed the prognostic parameters, such as OS, disease-specific survival (DSS), and progression-free interval (PFS), using the data of glioma patients from the TCGA in the Xiantao platform (https://www.xiantao.love/). These analyses were performed using the Cox regression and Kaplan–Meier methods. The low and high SLC31A1 gene expression groups were separated by the median value. We used the Wilcoxon signed-rank sum test and logistic regression to analyze the relationship between clinical-pathological characteristics and SLC31A1 expression. We used a multivariate Cox regression model to investigate the effect of SLC31A1 gene expression on the likelihood of survival and other clinical variables. A p value less than 0.05 was set as the threshold for significance. The Cox regression model findings were combined with the independent prognostic variables obtained from the multivariate analysis, and the survival probabilities for 1, 3, and 5 years were projected using these data. The projected odds were compared to actual occurrences using calibration curves. The 45-degree line represented the most accurately predicted value.
Protein interaction analysis in silico
The STRING platform (https://string-db.org/) was used for protein correlation data analysis. It provides integrated and consolidated PPI (protein–protein interaction) data. After importing the SLC31A1 expression data into the STRING platform, we received information from the PPI network. The threshold was set at a confidence score greater than 0.7. To get a more accurate result of the SLC31A1-related genes, the p value was adjusted by the Benjamini–Hochberg method, only the genes with adjusted p value (padj) < 0.05 was considered significant.
Analysis of the immune cell infiltration in silico
The research of Bindea G et al. showed that different cell markers could characterize different types of immune cells (Bindea et al. 2013). We then investigated the tumor infiltrated 24 different types of immune cells using the ssGSEA method using the data from TCGA. The Spearman correlation algorithm was used to compare immune cell infiltration levels between high and low SLC31A1 gene expression. The link between SLC31A1 gene expression level and immune infiltration, the association between infiltrating levels of immune cells, and the values obtained in various SLC31A1 gene expression subgroups were analyzed in the module of the “Xiantao tool” based on the findings of immune infiltration, Xiantao tool Spearman correlation, and Wilcoxon signed-rank sum. A p value < 0.05 was considered statistically significant (*p < 0.05; **p < 0.01; ***p < 0.001; and ****p < 0.0001).
Gene correlation analysis
GEPIA (http://gepia.cancer-pku.cn/index.html) is a web platform providing information on 9736 cancer types and 8587 specimens derived from TCGA and GTEx. It usually focuses on the analysis of the RNA-seq findings. The Gene and Isoform classes each specify the types of the corresponding number of types of genes and isoforms, which total 60,498 and 198,619, respectively. An investigation was conducted in the GEPIA database to confirm the relationship between the expression of the SLC31A1 gene and various immune cell markers. The expression degree of the SLC31A1 gene is shown along the x-axis, whereas the expression of other relevant genes is shown on the y-axis. Furthermore, using data from TIMER (http://cistrome.org/TIMER/), we confirmed the expression of genes with a strong relationship to SLC31A1 gene expression in GEPIA. A p value < 0.05 was statistically significant (*p < 0.05; **p < 0.01; ***p < 0.001; and ****p < 0.0001).
Quantitative real-time polymerase chain reaction (qPCR)
The total mRNAs were extracted with TRIzol reagent (Ambion, REF:15,596,018); then, we reversed the mRNAs to cDNAs using the cDNA Synthesis SuperMix kit (cat. E047-01B). We used 2 × RealStar Green Fast Mixture qPCR kit (GenStar, Cat: A301-101) to analyze the expression of the SLC31A1 gene in different glioma cell lines, in which GAPDH was used as the housekeeping gene, and its reliability was determined by the computational program RefFinder (Xie et al. 2023). The primers are as follows:
SLC31A1
Forward: 5′-ATGGAACCATCCTTATGGAGACA-3′
Backward: 5′-GGAAGTAGCTTATGACCACCTGG-3′
GAPDH
Forward: 5′-TGACTTCAACAGCGACACCCA-3′
Backward: 5′-CACCCTGTTGCTGTAGCCAAA-3′
The thermal cycle condition for qPCR: 95 ℃, 10 min; 95 ℃, 15 s; 60 ℃, 60 s, 40cycles.
The experiment has been repeated three times.
Knockdown of SLC31A1 in glioma cell lines
The siRNA targeting SLC31A1 was ordered from the OBiO company. The sequences of siRNAs are shown behind. The glioma cell lines LN229 and U251 were evenly planted in 6-well plates (1 × 105/well) in Dulbecco’s Modified Eagle’s Medium (Gibco) supplemented with 10% fetal bovine serum (PAN, Germany) and 100 μL/mL penicillin and streptomycin and placed at 37 °C with 5% CO2. siRNA of 110 pmol was transfected into per well; the transfection buffer was purchased from PolyPlus (jetPRIME® transfection reagent). The transfection was performed as suggested in the user guidance. The transfect efficiency was examined by the qPCR method. The experiment has been repeated for three times.
SiRNA for human SLC31A1
Si126
Forward: 5′-GAUGCCUAUGACCUUCUACUUTT-3′
Backward: 5′-AAGUAGAAGGUCAUAGGCAUCTT-3′
Si282
Forward: 5′-GCGUAAGUCACAAGUCAGCAUTT-3′
Backward: 5′-AUGCUGACUUGUGACUUACGCTT-3′
Si504
Froward: 5′- CGGUACAGGAUACUUCCUCUUTT-3′
Backward: 5′-AAGAGGAAGUAUCCUGUACCGTT-3′
SiNC
Forward: 5′-UUCUCCGAACGUGUCACGUTT-3′
Backward: 5′-ACGUGACACGUUCGGAGAATT-3′
CCK-8 assay
One thousand cells were seeded in 96-well cell plates and added CCK-8 solution (Beyotime biotechnology: C0010) at 24, 48, 72, 96, and 120 h. Two hours later, the OD value at 450 nm was measured by the microplate reader (Thermo-Fisher Scientific). The CCK-8 assay has been repeated for three times.
Transwell assay
Cells in the logarithmic growth phase were adjusted to 2 × 104 cells/well of 200 uL medium (without serum) and plated into the upper chamber (Corning: 3422). The lower chamber was added with 750 μL of medium (with 10% FBS), and then incubate the chamber at 37 °C for 20 h. Then, the migrated cells were fixed by 4% paraformaldehyde (LABLEAD: P4500, 20 min) and visualized by the 0.1% crystal violet (Solarbio: G1063, 20 min). The cells not penetrated were gently wept off by cotton. After drying, cells in four random visual fields were counted under an inverted microscope (Leica, Wetzlar, Germany) to obtain the mean value of cells. For the accuracy of our results, the transwell assay has been repeated for three times.
Flow cytometry
Transfected cells were gently collected and adjusted to a cell concentration of 1 × 105 cells/195 uL conjugation buffer (C1062M, Beyotime). Cells were stained with 5 uL Annexin V-fluorescein isothiocyanate (FITC) and 10 uL propidium iodide (PI) at RT (room temperature) for 15 to 20 min. The FITC and PI both positive cells were considered apoptotic and detected using a flow cytometer (CytoFLEX, Beckman Coulter). The apoptosis rate was calculated by the percentage of apoptotic cells in the total cells. The experiment has been repeated for three times and statistically analyzed using the prism.
Statistical analysis
The statistical analysis was performed by the Prism v.8.0.2 software (GraphPad). Statistical significance was also determined using the prism. A p value < 0.05 was statistically significant (*p < 0.05; **p < 0.01; ***p < 0.001; and ****p < 0.0001).
Data availability
All data generated or analyzed during this study are included in this published article.
References
Abulaiti A et al (2023) Molecular subtypes based on PANoptosis-related genes and tumor microenvironment infiltration characteristics in lower-grade glioma. Funct Integr Genomics 23(2):84
Armstrong TS et al (2020) Glioma patient-reported outcome assessment in clinical care and research: a Response assessment in neuro-oncology collaborative report. Lancet Oncol 21(2):e97–e103
Bi J et al (2020) Altered cellular metabolism in gliomas - an emerging landscape of actionable co-dependency targets. Nat Rev Cancer 20(1):57–70
Bindea G et al (2013) Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39(4):782–795
D'Alessio A, Proietti G, Sica G, Scicchitano BM (2019) Pathological and molecular features of glioblastoma and its peritumoral tissue. Cancers (Basel) 11(4):469. https://doi.org/10.3390/cancers11040469
Du Y et al (2022) Cuproptosis patterns and tumor immune infiltration characterization in colorectal cancer. Front Genet 13:976007
Eckel-Passow JE et al (2015) Glioma Groups Based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med 372(26):2499–2508
Friebel E et al (2020) Single-cell mapping of human brain cancer reveals tumor-specific instruction of tissue-invading leukocytes. Cell 181(7):1626-1642.e20
Gordon S, Martinez FO (2010) Alternative activation of macrophages: mechanism and functions. Immunity 32(5):593–604
Hambardzumyan D, Gutmann DH, Kettenmann H (2016) The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci 19(1):20–27
Jenkins RB et al (2006) A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res 66(20):9852–9861
Jiang T et al (2021) Clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett 499:60–72
Klemm F et al (2020) Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune. Cells Cell 181(7):1643-1660 e17
Kong F et al (2023) Systematic pan-cancer analysis identifies SLC31A1 as a biomarker in multiple tumor types. BMC Med Genomics 16(1):61
Lewinska-Preis L et al (2011) Bioelements and mineral matter in human livers from the highly industrialized region of the Upper Silesia Coal Basin (Poland). Environ Geochem Health 33(6):595–611
Li L, Li L, Sun Q (2022a) High expression of cuproptosis-related SLC31A1 gene in relation to unfavorable outcome and deregulated immune cell infiltration in breast cancer: an analysis based on public databases. BMC Bioinformatics 23(1):350
Li X, Ma Z, Mei L (2022b) Cuproptosis-related gene SLC31A1 is a potential predictor for diagnosis, prognosis and therapeutic response of breast cancer. Am J Cancer Res 12(8):3561–3580
Li J et al (2023) Radiation induces IRAK1 expression to promote radioresistance by suppressing autophagic cell death via decreasing the ubiquitination of PRDX1 in glioma cells. Cell Death Dis 14(4):259
Li L, Leng W, Chen J, Li S, Lei B, Zhang H, Zhao H (2023) Identification of a copper metabolism-related gene signature for predicting prognosis and immune response in glioma. Cancer Med 12(8):10123–10137. https://doi.org/10.1002/cam4.5688
Liu T et al (2022) Ferroptosis, as the most enriched programmed cell death process in glioma, induces immunosuppression and immunotherapy resistance. Neuro Oncol 24(7):1113–1125
Lv H et al (2022) Comprehensive analysis of cuproptosis-related genes in immune infiltration and prognosis in melanoma. Front Pharmacol 13:930041
Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM (2000) M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol 164(12):6166–6173. https://doi.org/10.4049/jimmunol.164.12.6166
Mosser DM, Edwards JP (2008) Exploring the full spectrum of macrophage activation. Nat Rev Immunol 8(12):958–969
Ochocka N et al (2021) Single-cell RNA sequencing reveals functional heterogeneity of glioma-associated brain macrophages. Nat Commun 12(1):1151
Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS (2018) CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011-2015. Neuro Oncol 20(suppl_4):iv1–iv86. https://doi.org/10.1093/neuonc/noy131
Quail DF, Joyce JA (2017) The microenvironmental landscape of brain tumors. Cancer Cell 31(3):326–341
Roszer T (2015) Understanding the mysterious M2 macrophage through activation markers and effector mechanisms. Mediators Inflamm 2015:816460
Sasmita AO, Wong YP, Ling APK (2018) Biomarkers and therapeutic advances in glioblastoma multiforme. Asia Pac J Clin Oncol 14(1):40–51
Shimato S, Maier LM, Maier R, Bruce JN, Anderson RC, Anderson DE (2012) Profound tumor-specific Th2 bias in patients with malignant glioma. BMC Cancer 12:561. https://doi.org/10.1186/1471-2407-12-561
Song Q et al (2022) Cuproptosis scoring system to predict the clinical outcome and immune response in bladder cancer. Front Immunol 13:958368
Stark JM, Tibbitt CA, Coquet JM (2019) The metabolic requirements of Th2 Cell Differentiation. Front Immunol 10:2318
Tsvetkov P et al (2022) Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 375(6586):1254–1261
Vidyarthi A et al (2019) Predominance of M2 macrophages in gliomas leads to the suppression of local and systemic immunity. Cancer Immunol Immunother 68(12):1995–2004
Walker JA, McKenzie ANJ (2018) T(H)2 cell development and function. Nat Rev Immunol 18(2):121–133
Wang T et al (2022a) Cuproptosis-related gene FDX1 expression correlates with the prognosis and tumor immune microenvironment in clear cell renal cell carcinoma. Front Immunol 13:999823
Wang X et al (2022b) Exploration and functionalization of M1-macrophage extracellular vesicles for effective accumulation in glioblastoma and strong synergistic therapeutic effects. Signal Transduct Target Ther 7(1):74
Wang G et al (2022c) Cuproptosis regulator-mediated patterns associated with immune in fi ltration features and construction of cuproptosis-related signatures to guide immunotherapy. Front Immunol 13:945516
Xie F, Wang J, Zhang B (2023) RefFinder: a web-based tool for comprehensively analyzing and identifying reference genes. Funct Integr Genomics 23(2):125
Xiong W et al (2022) Glioblastoma: two immune subtypes under the surface of the cold tumor. Aging 14(10):4357–4375
Xu J et al (2021) Hypoxic glioma-derived exosomes promote M2-like macrophage polarization by enhancing autophagy induction. Cell Death Dis 12(4):373
Yu Z et al (2023) Integration of clinical and spatial data to explore lipid metabolism-related genes for predicting prognosis and immune microenvironment in gliomas. Funct Integr Genomics 23(2):82
Yunna C et al (2020) Macrophage M1/M2 polarization. Eur J Pharmacol 877:173090
Zhang WT et al (2022a) A Novel cuprotosis-related gene FDX1 signature for overall survival prediction in clear cell renal cell carcinoma patients. Biomed Res Int 2022:9196540
Zhang G, Sun J, Zhang X (2022b) A novel Cuproptosis-related LncRNA signature to predict prognosis in hepatocellular carcinoma. Sci Rep 12(1):11325
Zhou B, Gitschier J (1997) hCTR1: a human gene for copper uptake identified by complementation in yeast. Proc Natl Acad Sci USA 94(14):7481–7486. https://doi.org/10.1073/pnas.94.14.7481
Zhu X, Zhu J (2020) CD4 T helper cell subsets and related human immunological disorders. Int J Mol Sci 21(21):8011. https://doi.org/10.3390/ijms21218011
Funding
This study was supported by the National Key R&D Program (2020YFC2004706).
Author information
Authors and Affiliations
Contributions
Jun Wang and Shenglun Li contributed the main idea and contributed equally to the main part of this article. Hongwei Zhang is the corresponding author of this research and he offered the funding and guidance for the study design. Yuduo Guo, Yujia Chen, and Weihai Ning offered technical support for data processing. Chao Zhao and Jingjing Yang completed the part of the experimental test. All authors contributed to the writing and revisions.
Corresponding author
Ethics declarations
Ethical approval
This investigation was carried out using online databases and cell lines; no ethical approval is needed in this study.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, J., Li, S., Guo, Y. et al. Cuproptosis-related gene SLC31A1 expression correlates with the prognosis and tumor immune microenvironment in glioma. Funct Integr Genomics 23, 279 (2023). https://doi.org/10.1007/s10142-023-01210-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10142-023-01210-0