Abstract
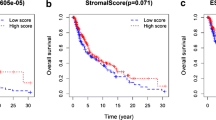
The tumor microenvironment (TME) dynamically regulates cancer progression and affects clinical outcomes. This study aimed to identify molecular subtypes and construct a prognostic risk model based on TME-related signatures in skin cutaneous melanoma (SKCM) patients. We categorized SKCM patients based on transcriptome data of SKCM from The Cancer Genome Atlas (TCGA) database and 29 TME-related gene signatures. Differentially expressed genes were identified using univariate Cox regression and Lasso regression analysis, which were used for risk model construction. The robustness of this model was validated in independent external cohorts. Genetic landscape alterations, immune characteristics, and responsiveness to immunotherapy/chemotherapy were evaluated. Three TME-related subtypes were identified, and subtype C3 exhibited the most favorable prognosis, had enriched immune-related pathways, and possessed more infiltration of T_cells_CD8, T_cells_CD4_memory_activated, and Macrophages_M1 but a lower TumorPurity, whereas Macrophages_M2 were increased in subtype C1 and subtype C2. Subtype C1 was more sensitive to Cisplatin, subtype C2 was more sensitive to Temozolomide, and subtype C3 was more sensitive to Paclitaxel; 8 TME-related genes (NOTCH3, HEYL, ZNF703, ABCC2, PAEP, CCL8, HAPLN3, and HPDL) were screened for risk model construction. High-risk patients had dismal prognosis with good prediction performance. Moreover, low-risk patients were more sensitive to Paclitaxel and Temozolomide, whereas high-risk patients were more sensitive to Cisplatin. This risk model had robustness in predicting prognosis in SKCM patients. The results facilitate the understanding of TME-related genes in SKCM and provide a TME-related genes-based predictive model in prognosis and direction of personalized options for SKCM patients.











Similar content being viewed by others
Data availability
The data supporting the conclusions of this article can be obtained in The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/) database and the Gene-Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) database.
References
Akbani R, Akdemir KC, Aksoy BA, Albert M, Ally A, Amin SB et al (2015) Genomic classification of cutaneous melanoma. Cell 161(7):1681–1696
Alamolhodaei NS, Rashidpour H, Gharaee ME, Behravan J, Mosaffa F (2020) Overexpression of ABCC2 and NF-Κβ/p65 with reduction in cisplatin and 4OH-Tamoxifen sensitivity in MCF-7 breast cancer cells: the influence of TNF-α. Pharmaceutical Sciences 26(2):150–158
Auslander N, Zhang G, Lee JS, Frederick DT, Miao B, Moll T et al (2018) Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat Med 24(10):1545–1549
Bagaev A, Kotlov N, Nomie K, Svekolkin V, Gafurov A, Isaeva O et al (2021) Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer cell 39(6):845–65.e7
Blanche P. (2015) TimeROC: time-dependent ROC curve and AUC for censored survival data. R package version 2
Chen H-y, Xu L, Li L-f, Liu X-x, Gao J-x, Bai Y-r (2018) Inhibiting the CD8+ T cell infiltration in the tumor microenvironment after radiotherapy is an important mechanism of radioresistance. Scientific reports 8(1):1–10
Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA (2018) Profiling tumor infiltrating immune cells with CIBERSORT. Springer, Cancer systems biology, pp 243–259
Chi H, Jiang P, Xu K, Zhao Y, Song B, Peng G et al (2022a) A novel anoikis-related gene signature predicts prognosis in patients with head and neck squamous cell carcinoma and reveals immune infiltration. Front Genet 13:984273
Chi H, Xie X, Yan Y, Peng G, Strohmer DF, Lai G et al (2022b) Natural killer cell-related prognosis signature characterizes immune landscape and predicts prognosis of HNSCC. Front Immunol 13:1018685
Chi H, Peng G, Yang J, Zhang J, Song G, Xie X et al (2022c) Machine learning to construct sphingolipid metabolism genes signature to characterize the immune landscape and prognosis of patients with uveal melanoma. Front Endocrinol 13:1056310
Chi H, Yang J, Peng G, Zhang J, Song G, Xie X et al (2023) Circadian rhythm-related genes index: a predictor for HNSCC prognosis, immunotherapy efficacy, and chemosensitivity. Front Immunol 14:1091218
Chi H, Peng G, Wang R, Yang F, Xie X, Zhang J, et al. (2022) Cuprotosis programmed-cell-death-related lncRNA signature predicts prognosis and immune landscape in PAAD patients. Cells 11 21
Cursons J, Souza-Fonseca-Guimaraes F, Foroutan M, Anderson A, Hollande F, Hediyeh-Zadeh S et al (2019) A Gene signature predicting natural killer cell infiltration and improved survival in melanoma patientsa gene signature for NK infiltration and melanoma survival. Cancer Immunol Res 7(7):1162–1174
Danilova L, Ho WJ, Zhu Q, Vithayathil T, De Jesus-Acosta A, Azad NS et al (2019) Programmed cell death ligand-1 (PD-L1) and CD8 expression profiling identify an immunologic subtype of pancreatic ductal adenocarcinomas with favorable survival. Cancer Immunol Res 7(6):886–895
Dizier B, Callegaro A, Debois M, Dreno B, Hersey P, Gogas HJ et al (2020) A Th1/IFNγ gene signature is prognostic in the adjuvant setting of resectable high-risk melanoma but not in non–small cell lung cancerprognostic biomarkers in metastatic melanoma and NSCLC. Clin Cancer Res 26(7):1725–1735
Geeleher P, Cox N, Huang RS (2014) pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE 9(9):e107468
Gu Z (2022) Complex Heatmap Visualization Imeta 1(3):e43
Hänzelmann S, Castelo R, Guinney J (2013) GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14(1):1–15
Hastie T, Qian J,Tay K. An introduction to glmnet. CRAN R Repositary
Hou Y-C, Chao Y-J, Hsieh M-H, Tung H-L, Wang H-C, Shan Y-S (2019) Low CD8+ T cell infiltration and high PD-L1 expression are associated with level of CD44+/CD133+ cancer stem cells and predict an unfavorable prognosis in pancreatic cancer. Cancers 11(4):541
Hu J, Hu W, Yang Y. (2022) Hypoxia Confers Tumor with a higher immune infiltration but lower mutation burden in gastrointestinal cancer. Journal of oncology
Huang K, Luo W, Fang J, Yu C, Liu G, Yuan X, et al. (2022) Notch3 signaling promotes colorectal tumor growth by enhancing immunosuppressive cells infiltration in the microenvironment
Jayasingam SD, Citartan M, Thang TH, Mat Zin AA, Ang KC, Ch’ng ES (2020) Evaluating the polarization of tumor-associated macrophages into M1 and M2 phenotypes in human cancer tissue: technicalities and challenges in routine clinical practice. Front Oncol 9:1512
Jiang N, Zhang X, Chen Q, Kantawong F, Wan S, Liu J, et al. (2022) Identification of a mitochondria-related gene signature to predict the prognosis in AML. Frontiers in oncology 12
Jin W, Yang Q, Chi H, Wei K, Zhang P, Zhao G et al (2022) Ensemble deep learning enhanced with self-attention for predicting immunotherapeutic responses to cancers. Front Immunol 13:1025330
Kasakovski D, Skrygan M, Gambichler T, Susok L (2021) Advances in Targeting Cutaneous Melanoma Cancers 13(9):2090
Kassambara A. 2019 ggcorrplot: visualization of a correlation matrix using ggplot2 (0.1. 3)[Computer software]
Kassambara A, Kosinski M, Biecek P, Fabian S. (2017) survminer: drawing survival curves using ‘ggplot2’. R package version 03 1
Liang Z, Pan L, Shi J, Zhang L (2022) C1QA, C1QB, and GZMB are novel prognostic biomarkers of skin cutaneous melanoma relating tumor microenvironment. Sci Rep 12(1):1–15
Maimela NR, Liu S, Zhang Y (2019) Fates of CD8+ T cells in tumor microenvironment. Comput Struct Biotechnol J 17:1–13
Mayakonda A, Lin D-C, Assenov Y, Plass C, Koeffler HP (2018) Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 28(11):1747–1756
Mei Y, Chen M-JM, Liang H, Ma L (2021) A four-gene signature predicts survival and anti-CTLA4 immunotherapeutic responses based on immune classification of melanoma. Communications biology 4(1):1–12
Meurette O, Mehlen P (2018) Notch signaling in the tumor microenvironment. Cancer Cell 34(4):536–548
Najafi M, Goradel NH, Farhood B, Salehi E, Solhjoo S, Toolee H et al (2019) Tumor microenvironment: interactions and therapy. J Cell Physiol 234(5):5700–5721
Novotný J, Strnadová K, Dvořánková B, Kocourková Š, Jakša R, Dundr P et al (2020) Single-cell RNA sequencing unravels heterogeneity of the stromal niche in cutaneous melanoma heterogeneous spheroids. Cancers 12(11):3324
Peng G, Chi H, Gao X, Zhang J, Song G, Xie X et al (2022) Identification and validation of neurotrophic factor-related genes signature in HNSCC to predict survival and immune landscapes. Front Genet 13:1010044
Ritchie ME,Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. (2015) Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research 43 7 e47-e
Santos P, I, van Doorn R, Caspers PJ, Bakker Schut TC, Barroso EM, Nijsten TE, et al (2018) Improving clinical diagnosis of early-stage cutaneous melanoma based on Raman spectroscopy. British journal of cancer 119(11):1339–46
Song B, Chi H, Peng G, Song Y, Cui Z, Zhu Y et al (2022a) Characterization of coagulation-related gene signature to predict prognosis and tumor immune microenvironment in skin cutaneous melanoma. Front Oncol 12:975255
Song B, Wu P, Liang Z, Wang J, Zheng Y, Wang Y et al (2022b) A novel necroptosis-related gene signature in skin cutaneous melanoma prognosis and tumor microenvironment. Front Genet 13:917007
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A et al (2020) Global cancer statistics 2020 GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers a cancers in 185 countries. CA a cancer journal for clinicians 71(3):209–49
Thrane K, Eriksson H, Maaskola J, Hansson J, Lundeberg J (2018) Spatially resolved transcriptomics enables dissection of genetic heterogeneity in stage III cutaneous malignant melanoma. Can Res 78(20):5970–5979
Wilkerson M,Waltman P, Wilkerson MM. 2013 Package ‘ConsensusClusterPlus’
Wu H, Huang S, Zhuang W, Qiao G (2021) Prognostic significance of eight immune-related genes on survival in patients with lung squamous cell carcinoma. Biomark Med 15(4):295–306
Xu B, Peng Z, Yan G, Wang N, Chen M, Yao X, et al. (2021) Establishment and validation of a genetic label associated with M2 Macrophage infiltration to predict survival in colon cancer patients and assist immunotherapy
Yang P, Chen W, Xu H, Yang J, Jiang J, Jiang Y et al (2021) Correlation of CCL8 expression with immune cell infiltration of skin cutaneous melanoma: potential as a prognostic indicator and therapeutic pathway. Cancer Cell Int 21(1):1–11
Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W et al (2013) Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 4(1):1–11
Yu G, Wang L-G, Han Y, He Q-Y. (2012) clusterProfiler an R package for comparing biological themes among gene clusters Omics a journal of integrative biology 16 5 284-7
Zhang Y, Zhang Z (2020) The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol 17(8):807–821
Zhao Y, Wei K, Chi H, Xia Z, Li X (2022) IL-7: a promising adjuvant ensuring effective T cell responses and memory in combination with cancer vaccines? Front Immunol 13:1022808
Zhao S, Chi H, Yang Q, Chen S, Wu C, Lai G et al (2023) Identification and validation of neurotrophic factor-related gene signatures in glioblastoma and Parkinson’s disease. Front Immunol 14:1090040
Zhou S, Sun Y, Chen T, Wang J, He J, Lyu J et al (2021) The landscape of the tumor microenvironment in skin cutaneous melanoma reveals a prognostic and immunotherapeutically relevant gene signature. Frontiers in Cell and Developmental Biology 9:739594
Zhu G, Su H, Johnson CH, Khan SA, Kluger H, Lu L (2021) Intratumour microbiome associated with the infiltration of cytotoxic CD8+ T cells and patient survival in cutaneous melanoma. Eur J Cancer 151:25–34
Funding
This study was funded by Xiang’an Hospital of Xiamen University and the Guidance in Medical and Health Program of Xiamen, China (3502Z20224ZD1148).
Author information
Authors and Affiliations
Contributions
All authors wrote, revised, and approved the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
The study was conducted in compliance with the ethical principles of the Declaration of Helsinki and its later amendments. The Ethics Committee of Xiang’an Hospital of Xiamen University approved our study protocol (approval no. XAHLL2022046).
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lian, W., Zheng, X. Identification and validation of TME-related signatures to predict prognosis and response to anti-tumor therapies in skin cutaneous melanoma. Funct Integr Genomics 23, 153 (2023). https://doi.org/10.1007/s10142-023-01051-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10142-023-01051-x