Abstract
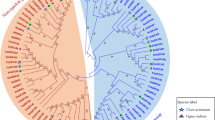
GF14 proteins are a family of conserved proteins involved in many cellular processes including transport, growth, metabolism, and stress response. However, only few reports are available regarding the 14-3-3 genes in potato. In this study, twelve 14-3-3 genes were detected in the potato genome. Based on their phylogenetic relationships, the StGF14 family members were categorized into two classes. Gene expression analysis demonstrated that StGF14h, StGF14a, and StGF14k had the highest gene expression, induced by abiotic and biotic stresses in all three tissues. The number of exons in 14-3-3 genes ranged from four to seven and most of these genes in the same subfamily had similar exon-intron patterns. The results of our study showed that the conserved motifs are similar in most of the proteins in each group. The intron-exon patterns and the composition of conserved motifs validated the 14-3-3 gene phylogenetic classification. According to the genome distribution results, 14-3-3 genes were located unevenly on the 12 Solanum tuberosum chromosomes. We find out 97 orthologous gene pairs between potato and Arabidopsis as well as 15 paralogous genes among potato genomes. Our results showed that GF-14 genes have an effective role in functional and molecular mechanisms in response to environmental stresses.











Similar content being viewed by others
References
Bihn EA, Paul AL, Wang SW, Erdos GW, Ferl RJ (1997) Localization of 14-3-3 proteins in the nuclei of Arabidopsis and maize. Plant J 12:1439–1445
Brandt J, Thordal-Christensen H, Vad K, Gregersen PL, Collinge DB (1992) A pathogen-induced gene of barley encodes a protein showing high similarity to a protein kinase regulator. Plant J 2:815–820
Bunney TD, van Walraven HS, de Boer AH (2001) 14-3-3 protein is a regulator of the mitochondrial and chloroplast ATP synthase. P N A S 98:4249–4254
Cao H, Xu Y, Yuan L, Bian Y, Wang L, Zhen S, Hu Y, Yan Y (2016) Molecular characterization of the 14-3-3 gene family in Brachypodium distachyon L. reveals high evolutionary conservation and diverse responses to abiotic stresses. Front Plant Sci 7:1099
Chen Z, Fu H, Liu D, Chang PF, Narasimhan M, Ferl R, Hasegawa PM, Bressan RA (1994) A NaCl-regulated plant gene encoding a brain protein homolog that activates ADP ribosyltransferase and inhibits protein kinase C. Plant J 6:729–740
Chen F, Li Q, Sun L, He Z (2006) The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress. DNA Res 13:53–63
Cheng C, Wang Y, Chai F, Li S, Xin H, Liang Z (2018) Genome-wide identification and characterization of the 14–3-3 family in Vitis vinifera L. during berry development and cold-and heat-stress response. BMC Genom 19:1–4
Faris JD, Li WL, Liu DJ, Chen PD, Gill BS (1999) Candidate gene analysis of quantitative disease resistance in wheat. Theor Appl Genet 98:219–225
Finnie C, Andersen CH, Borch J, Gjetting S, Christensen AB, De Boer AH, Thordal-Christensen H, Collinge DB (2002) Do 14-3-3 proteins and plasma membrane H+-ATPases interact in the barley epidermis in response to the barley powdery mildew fungus? Plant Mol Biol 49:137–147
Gao WJ, Chen Q, Zhao JY, Wang P, Li DL, Zheng K, Long YL, Jiao Y, Wang YX, Geng SW, Su XN (2020) Genome-wide identification and expression analysis of the 14-3-3 gene family in cotton (Gossypium hirsutum L.). PeerJ. 7:7950
Hajibarat Z, Saidi A, Zeinalabedini M, Gorji AM, Ghaffari MR, Shariati V, Ahmadvand R (2022) Genome-wide identification of StU-box gene family and assessment of their expression in developmental stages of Solanum tuberosum. J Genet Eng Biotechnol 20:1–21
Hill MK, Lyon KJ, Lyon BR (1999) Identification of disease response genes expressed in Gossypium hirsutum upon infection with the wilt pathogen Verticillium dahliae. Plant Mol Biol 40:289–296
Li X, Wang QJ, Pan N, Lee S, Zhao Y, Chait BT, Yue Z (2011) Phosphorylation-dependent 14-3-3 binding to LRRK2 is impaired by common mutations of familial Parkinson’s disease. PLoS One 6:17153
Li MY, Xu BY, Liu JH, Yang XL, Zhang JB, Jia CH, Ren LC, Jin ZQ (2012) Identification and expression analysis of four 14-3-3 genes during fruit ripening in banana (Musa acuminata L. AAA group, cv. Brazilian). Plant Cell Rep 31:369–378
Lozano-Durán R, Robatzek S (2015) 14-3-3 proteins in plant-pathogen interactions. Mol Plant-Microbe Interact 28:511–518
Malerba M, Cerana R, Crosti P (2004) Comparison between the effects of fusicoccin, Tunicamycin, and Brefeldin A on programmed cell death of cultured sycamore (Acer pseudoplatanus L.) cells. Protoplasma 224:61–70
Qin C, Cheng L, Shen J, Zhang Y, Cao H, Lu D, Shen C (2016) Genome-wide identification and expression analysis of the 14-3-3 family genes in Medicago truncatula. Front Plant Sci 7:320
Roberts MR (2003) 14-3-3 proteins find new partners in plant cell signalling. Trends Plant Sci 8:218–223
Roberts MR, Bowles DJ (1999) Fusicoccin, 14-3-3 proteins, and defense responses in tomato plants. Plant Physiol 119:1243–1250
Rosenquist M, Alsterfjord M, Larsson C, Sommarin M (2001) Data mining the Arabidopsis genome reveals fifteen 14-3-3 genes. Expression is demonstrated for two out of five novel genes. Plant Physiol 127:142–149
Roth C, Liberles DA (2006) A systematic search for positive selection in higher plants (Embryophytes). BMC plant Biol 6:1–11
Saidi A, Hajibarat Z (2020a) Application of next generation sequencing, GWAS, RNA seq, WGRS, for genetic improvement of potato (Solanum tuberosum L) under drought stress. Biocatal Agric Biotechnol 29:101801
Saidi A, Hajibarat Z. 2020b. Computational analysis for characterization and evaluation of pentatricopeptide repeat-containing protein (PPR) in Arabidopsis thaliana. Polish J Nat Sci. 35.
Saidi A, Hajibarat Z, Hajibarat Z (2021a) Phylogeny, gene structure and GATA genes expression in different tissues of Solanaceae species. Biocatal Agric Biotechnol 35:102015
Saidi A, Hajibarat Z (2021b) Approaches for developing molecular markers associated with virus resistances in potato (Solanum tuberosum). J Plant Dis Prot 18:1–4
Seehaus K, Tenhaken R (1998) Cloning of genes by mRNA differential display induced during the hypersensitive reaction of soybean after inoculation with Pseudomonas syringae pv. glycinea. Plant Mol Biol 38:1225–1234
Sehnke PC, Chung H-J, Wu K, Ferl RJ (2001) Regulation of starch accumulation by granule-associated plant 14-3-3 proteins. P Natl Acad Sci USA 98:765–770
Sun G, Xie F, Zhang B (2011) Transcriptome-wide identification and stress properties of the 14-3-3 gene family in cotton (Gossypium hirsutum L.). Funct Integat Genom. 11:627–636
Tian F, Wang T, Xie Y, Zhang J, Hu J (2015) Genome-wide identification, classification, and expression analysis of 14-3-3 gene family in Populus. PLoS One 10:1–2210
Vatansever R, Ozyigit II, Filiz E, Gozukara N (2017) Genome-wide exploration of silicon (Si) transporter genes, Lsi1 and Lsi2 in plants; insights into Si-accumulation status/capacity of plants. Biometals 30:185–200
Wang J, Sun P, Li Y, Liu Y, Yu J, Ma X, Sun S, Yang N, Xia R, Lei T. (2017). Hierarchically aligning 10 legume Wang Y, Ling L, Jiang Z, Tan W, Liu Z, Wu L, Zhao Y, Xia S, Ma J, Wang G, Li W. Genome-wide identification and expression analysis of the 14-3-3 gene family in soybean (Glycine max). Peer J. 2019 7: 7950.
Yan J, Wang J, Zhang H (2002) An ankyrin repeat-containing protein plays a role in both disease resistance and antioxidation metabolism. Plant J 29:193–202
Acknowledgements
The authors are grateful for receiving assistance from the Shahid Beheshti University, Tehran, Iran.
Author information
Authors and Affiliations
Contributions
ZH helped in carrying out the experiment and writing the MS. AS helped in analysis and editing of the manuscript. ZH helped in writing of the manuscript. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hajibarat, Z., Saidi, A. & Hajibarat, Z. Genome-wide identification of 14-3-3 gene family and characterization of their expression in developmental stages of Solanum tuberosum under multiple biotic and abiotic stress conditions. Funct Integr Genomics 22, 1377–1390 (2022). https://doi.org/10.1007/s10142-022-00895-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-022-00895-z