Abstract
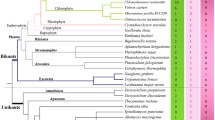
Myeloblastosis (MYB) family, the largest plant transcription factor family, has been subcategorised based on the number and type of repeats in the MYB domain. In spite of several reports, evolution of MYB genes and repeats remains enigmatic. Brassicaceae members are endowed with complex genomes, including dysploidy because of its unique history with multiple rounds of polyploidisation, genomic fractionations and rearrangements. The present study is an attempt to gain insights into the complexities of MYB family diversity, understand impacts of genome evolution on gene families and develop an evolutionary framework to understand the origin of various subcategories of MYB gene family. We identified and analysed 1129 MYBs that included 1R-, 2R-, 3R- and atypical-MYBs across sixteen species representing protists, fungi, animals and plants and exclude MYB identified from Brassicaceae except Arabidopsis thaliana; in addition, a total of 1137 2R-MYB genes from six Brassicaceae species were also analysed. Comparative analysis revealed predominance of 1R-MYBs in protists, fungi, animals and lower plants. Phylogenetic reconstruction and analysis of selection pressure suggested ancestral nature of R1-type repeat containing 1R-MYBs that might have undergone intragenic duplication to form multi-repeat MYBs. Distinct differences in gene structure between 1R-MYB and 2R-MYBs were observed regarding intron number, the ratio of gene length to coding DNA sequence (CDS) length and the length of exons encoding the MYB domain. Conserved as well as novel and lineage-specific intron phases were identified. Analyses of physicochemical properties revealed drastic differences indicating functional diversification in MYBs. Phylogenetic reconstruction of 1R- and 2R-MYB genes revealed a shared structure–function relationship in clades which was supported when transcriptome data was analysed in silico. Comparative genomics to study distribution pattern and mapping of 2R-MYBs revealed congruency and greater degree of synteny and collinearity among closely related species. Micro-synteny analysis of genomic segments revealed high conservation of genes that are immediately flanking the surrounding tandemly organised 2R-MYBs along with instances of local duplication, reorganisations and genome fractionation. In summary, polyploidy, dysploidy, reshuffling and genome fractionation were found to cause loss or gain of 2R-MYB genes. The findings need to be supported with functional validation to understand gene structure–function relationship along the evolutionary lineage and adaptive strategies based on comparative functional genomics in plants.











Similar content being viewed by others
References
Albert VA, Barbazuk WB, Depamphilis CW, Der JP, Leebens-Mack J, Ma H, Palmer JD, Rounsley S, Sankoff D, Schuster SC, Soltis DE (2013) The Amborella genome and the evolution of flowering plants. Science 342(6165):1241089
Anand S, Lal M, Das S (2019) Comparative genomics reveals origin of MIR159A-MIR159B paralogy, and complexities of PTGS interaction between miR159 and target GA-MYBs in Brassicaceae. Mol Genet Genomics 294(3):693–714
Anisimova M, Gascuel O (2006) Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol 55(4):539–552
Babenko VN, Rogozin IB, Mekhedov SL, Koonin EV (2004) Prevalence of intron gain over intron loss in the evolution of paralogous gene families. Nucl Acids Res 32(12):3724–3733
Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, Wishart DS (2016) Heatmapper: web-enabled heat mapping for all. Nucl Acid Res 44(W1):W147-153
Ballesteros ML, Bolle C, Lois LM, Moore JM, Vielle-Calzada JP, Grossniklaus U, Chua NH (2001) LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev 15(19):2613–2625
Banks JA, Nishiyama T, Hasebe M, Bowman JL, Gribskov M, DePamphilis C, Albert VA, Aono N, Aoyama T, Ambrose BA, Ashton NW (2011) The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332(6032):960–963
Bhattacharya R, Rose PW, Burley SK, Prlić A (2017) Impact of genetic variation on three-dimensional structure and function of proteins. PLoS One 12(3): e0171355
Bowen AJ, Gonzalez D, Mullins JG, Bhatt AM, Martinez A, Conlan RS (2010) PAH-domain-specific interactions of the Arabidopsis transcription coregulator SIN3-LIKE1 (SNL1) with telomere-binding protein 1 and ALWAYS EARLY2 Myb-DNA binding factors. J Mol Biol 395(5):937–949
Bowman JL, Kohchi T, Yamato KT, Jenkins J, Shu S, Ishizaki K, Yamaoka S, Nishihama R, Nakamura Y, Berger F, Adam C (2017) Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171(2):287–304
Bray N, Dubchak I, Pachter L (2003) AVID: A global alignment program. Genome Res 13(1):97–102
Cai B, Li CH, Huang J (2014) Systematic identification of cell-wall related genes in Populus based on analysis of functional modules in co-expression network. PloS one 9(4):e95176
Carre IA, Kim JY (2002) MYB transcription factor in the Arabidopsis circadian clock. J Exp Bot 53(374):1551–1557
Chahtane H, Vachon G, Le Masson M, Thévenon E, Périgon S, Mihajlovic N, Kalinina A, Michard R, Moyroud E, Monniaux M, Sayou C (2013) A variant of LEAFY reveals its capacity to stimulate meristem development by inducing RAX1. Plant J 74(4):678–689
Chang X, Xie S, Wei L, Lu Z, Chen ZH, Chen F, Lai Z, Lin Z, Zhang L (2020) Origins and stepwise expansion of R2R3-MYB transcription factors for the terrestrial adaptation of plants. Front Plant Sci 11:2086
Chen J, Glémin S, Lascoux M (2017) Genetic diversity and the efficacy of purifying selection across plant and animal species. Mol Biol Evol 34(6):1417–1428
Chen Y, Chen Z, Kang J, Kang D, Gu H, Qin G (2013) AtMYB14 regulates cold tolerance in Arabidopsis. Plant Mol Biol Report 31(1):87–97
Cheng H, Song S, Xiao L, Soo HM, Cheng Z, Xie D, Peng J (2009) Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genetics 5(3):e1000440
Cheong WH, Tan YC, Yap SJ, Ng KP (2015) ClicO FS: an interactive web-based service of Circos. Bioinformatics 31(22):3685–3687
Cohen H, Fedyuk V, Wang C, Wu S, Aharoni A (2020) SUBERMAN regulates developmental suberization of the Arabidopsis root endodermis. Plant J 102(3):431–447
Couronne O, Poliakov A, Bray N, Ishkhanov T, Ryaboy D, Rubin E, Pachter L, Dubchak I (2003) Strategies and tools for whole-genome alignments. Genome Res 13(1):73–80
Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14(6):1188–1190
Dai X, Zhou L, Zhang W, Cai L, Guo H, Tian H, Wang S (2016) A single amino acid substitution in the R3 domain of GLABRA1 leads to inhibition of trichome formation in Arabidopsis without affecting its interaction with GLABRA3. Plant, Cell Environ 39(4):897–907
Dangwal M, Das S (2018) Identification and analysis of OVATE family members from genome of the early land plants provide insights into evolutionary history of OFP family and function. J Mol Evol 86:511–530
Delk NA, Johnson KA, Chowdhury NI, Braam J (2005) CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in responses to abscisic acid, daylength, and ion stress. Plant Physiol 139(1):240–253
Ding Y, Zhou Q, Wang W (2012) Origins of new genes and evolution of their novel functions. Annu Rev Ecol Evol Syst 43(1):345–363
Du H, Liang Z, Zhao S, Nan MG, Tran LS, Lu K, Huang YB, Li JN (2015) The evolutionary history of R2R3-MYB proteins across 50 eukaryotes: new insights into subfamily classification and expansion. Sci Rep 5(1):1–6
Du H, Zhang L, Liu L, Tang XF, Yang WJ, Wu YM, Huang YB, Tang YX (2009) Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry (mosc) 74(1):1–11
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15(10):573–581
Fang Q, Wang G, Mao H, Xu J, Wang Y, Hu H, He S, Tu J, Cheng C, Tian G, Wang X, Liu X, Zhang C, Luo K (2018) AtDIV2, an R-R-type MYB transcription factor of Arabidopsis, negatively regulates salt stress by modulating ABA signaling. Plant Cell Rep 37:1499–1511
Feng G, Burleigh JG, Braun EL, Barbazuk WB (2017) Evolution of the 3R-MYB Gene Family in Plants. Genome Biol Evol 9(4):1013–1029
Fernandez-Pozo N, Haas FB, Meyberg R, Ullrich KK, Hiss M, Perroud PF, Hanke S, Kratz V, Powell AF, Vesty EF, Daum CG, Zane M, Lipzen A, Sreedasyam A, Grimwood J, Coates JC, Barry K, Schmutz J, Mueller LA, Rensing SA (2020) PEATmoss (Physcomitrella Expression Atlas Tool): a unified gene expression atlas for the model plant Physcomitrella patens. Plant J 102(1):165–177
Ferrari C, Shivhare D, Hansen BO, Pasha A, Esteban E, Provart NJ, Kragler F, Fernie A, Tohge T, Mutwil M (2020) Expression atlas of Selaginella moellendorffii provides insights into the evolution of vasculature, secondary metabolism, and roots. Plant Cell 32(4):853–870
Fornale S, Lopez E, Salazar-Henao JE, Fernández-Nohales P, Rigau J, Caparros-Ruiz D (2014) AtMYB7, a new player in the regulation of UV-sunscreens in Arabidopsis thaliana. Plant Cell Physiol 55(3):507–516
Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Research 32(suppl_2):W273–279
Frerigmann H, Gigolashvili T (2014) MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol Plant 7(5):814–828
Geng P, Zhang S, Liu J, Zhao C, Wu J, Cao Y, Zhao Q (2020) MYB20, MYB42, MYB43, and MYB85 regulate phenylalanine and lignin biosynthesis during secondary cell wall formation. Plant Physiol 182(3):1272–1283
Glusman G, Rose PW, Prlić A, Dougherty J, Duarte JM, Hoffman AS, Barton GJ, Bendixen E, Bergquist T, Bock C, Brunk E, Buljan M, Burley SK, Cai B, Carter H, Gao J, Godzik A, Heuer M, Hicks M, Hrabe T, Karchin R, Leman JK, Lane L, Masica DL, Mooney SD, Moult J, Omenn GS, Pearl F, Pejaver V, Reynolds SM, Rokem A, Schwede T, Song S, Tilgner H, Valasatava Y, Zhang Y, Deutsch EW (2017) Mapping genetic variations to three-dimensional protein structures to enhance variant interpretation: a proposed framework. Genome Med 9(1):113
Gocal GF, Sheldon CC, Gubler F, Moritz T, Bagnall DJ, MacMillan CP, Li SF, Parish RW, Dennis ES, Weigel D, King RW (2001) GAMYB-like genes, flowering, and gibberellin signaling in Arabidopsis. Plant Physiol 127(4):1682–1693
Gonzalez A, Mendenhall J, Huo Y, Lloyd A (2009) TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev Biol 325(2):412–421
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59(3):307–21
Guo D, Zhang J, Wang X, Han X, Wei B, Wang J, Li B, Yu H, Huang Q, Gu H, Qu LJ (2015) The WRKY transcription factor WRKY71/EXB1 controls shoot branching by transcriptionally regulating RAX genes in Arabidopsis. Plant Cell 27(11):3112–3127
Hamaguchi A, Yamashino T, Koizumi N, Kiba T, Kojima M, Sakakibara H, Mizuno T (2008) A small subfamily of Arabidopsis RADIALIS-LIKE SANT/MYB genes: a link to HOOKLESS1-mediated signal transduction during early morphogenesis. Biosci Biotechnol Biochem 72(10):2687–2696
Hanaoka S, Nagadoi A, Yoshimura S, Aimoto S, Li B, de Lange T, Nishimura Y (2001) NMR structure of the hRap1 Myb motif reveals a canonical three-helix bundle lacking the positive surface charge typical of Myb DNA-binding domains. J Mol Biol 312(1):167–75
He Q, Jones DC, Li W, Xie F, Ma J, Sun R, Wang Q, Zhu S, Zhang B (2016) Genome-wide identification of R2R3-MYB genes and expression analyses during abiotic stress in Gossypium raimondii. Sci Rep 6(1):1–4
Higo A, Kawashima T, Borg M, Zhao M, López-Vidriero I, Sakayama H, Montgomery SA, Sekimoto H, Hackenberg D, Shimamura M, Nishiyama T (2018) Transcription factor DUO1 generated by neo-functionalization is associated with evolution of sperm differentiation in plants. Nat Commun 9(1):1–13
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31(8):1296–7
Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, Haberer G (2011) The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43(5):476–481
Jain A, Das S (2016) Synteny and comparative analysis of miRNA retention, conservation, and structure across Brassicaceae reveals lineage- and sub-genome-specific changes. Funct Integr Genomics 16(3):253–268
Jeong CY, Kim JH, Lee WJ, Jin JY, Kim J, Hong SW, Lee H (2018) AtMyb56 regulates anthocyanin levels via the modulation of AtGPT2 expression in response to sucrose in Arabidopsis. Mol Cells 41(4):351
Jiang C, Gu J, Chopra S, Gu X, Peterson T (2004) Ordered origin of the typical two- and three-repeat Myb genes. Gene 326:13–22
Jiang CK, Rao GY (2020) Insights into the diversification and evolution of R2R3-MYB transcription factors in plants. Plant Physiol 183(2):637–655
Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19(22):6150–6161
Jin JP, Tian F, Yang DC, Meng YQ, Kong L, Luo JC and Gao G (2017) PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Research 45(D1):D1040-D1045
Jung C, Kim YK, Oh NI, Shim JS, Seo JS, Do Choi Y, Nahm BH, Cheong JJ (2012) Quadruple 9-mer-based protein binding microarray analysis confirms AACnG as the consensus nucleotide sequence sufficient for the specific binding of AtMYB44. Mol Cells 34(6):531–537
Kagale S, Koh C, Nixon J, Bollina V, Clarke WE, Tuteja R, Spillane C, Robinson SJ, Links MG, Clarke C, Higgins EE (2014) The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat Commun 5(1):1–11
Katoh K, Rozewicki J, Yamada KD (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20(4):1160–1166
Keese PK, Gibbs A (1992) Origins of genes: “big bang” or continuous creation? Proc Natl Acad Sci 89(20):9489–9493
Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50(2):347–363
Kim SH, Kim HS, Bahk S, An J, Yoo Y, Kim JY, Chung WS (2017) Phosphorylation of the transcriptional repressor MYB15 by mitogen-activated protein kinase 6 is required for freezing tolerance in Arabidopsis. Nucleic Acids Res 45(11):6613–6627
Kim MK, Kim WT (2018) Telomere structure, function, and maintenance in plants. J Plant Biol 61:131–136
Klempnauer KH, Gonda TJ, Bishop JM (1982) Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: the architecture of a transduced oncogene. Cell 31(2):453–463
Konig P, Fairall L, Rhodes D (1998) Sequence-specific DNA recognition by the myb-like domain of the human telomere binding protein TRF1: a model for the protein-DNA complex. Nucleic Acids Res 26(7):1731–1740
Konig P, Rhodes D (1997) Recognition of telomeric DNA. Trends Biochem Sci 22(2):43–47
Koonin EV (2005) Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet 39:309–338
Koonin EV (2006) The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate? Biol Direct 1:22. https://doi.org/10.1186/1745-6150-1-22
Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C, Smeekens S (1998) Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J 16(2):263–276
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549
Lang D, Ullrich KK, Murat F, Fuchs J, Jenkins J, Haas FB, Piednoel M, Gundlach H, Van Bel M, Meyberg R, Vives C (2018) The Physcomitrella patens chromosome-scale assembly reveals moss genome structure and evolution. Plant J 93(3):515–533
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–8
Lashbrooke J, Cohen H, Levy-Samocha D, Tzfadia O, Panizel I, Zeisler V, Aharoni A (2016) MYB107 and MYB9 homologs regulate suberin deposition in angiosperms. Plant Cell 28(9):2097–2116
Lee DK, Geisler M, Springer PS (2009) LATERAL ORGAN FUSION1 and LATERAL ORGAN FUSION2 function in lateral organ separation and axillary meristem formation in Arabidopsis. Development 136(14):2423–2432
Lee SB, Kim HU, Suh MC (2016) MYB94 and MYB96 additively activate cuticular wax biosynthesis in Arabidopsis. Plant Cell Physiol 57(11):2300–2311
Lee TH, Tang H, Wang X, Paterson AH (2012) PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res 41(D1):D1152–D1158
Lefort V, Longueville JE, Gascuel O. SMS: smart model selection in PhyML (2017) Molecular Biology and Evolution 34(9):2422–4
Liang G, He H, Li Y, Ai Q, Yu D (2014) MYB82 functions in regulation of trichome development in Arabidopsis. J Exp Bot 65(12):3215–3223
Liang Y, Tan ZM, Zhu L, Niu QK, Zhou JJ, Li M, Ye D (2013) MYB97, MYB101 and MYB120 function as male factors that control pollen tube-synergid interaction in Arabidopsis thaliana fertilization. PLoS genetics 9(11):e1003933
Lin Z, Yin K, Zhu D, Chen Z, Gu H, Qu LJ (2007) AtCDC5 regulates the G2 to M transition of the cell cycle and is critical for the function of Arabidopsis shoot apical meristem. Cell Res 17(9):815–828
Lipsick JS (1996) One billion years of Myb. Oncogene 13(2):223–235
Liu L, Zhang J, Adrian J, Gissot L, Coupland G, Yu D, Turck F (2014) Elevated levels of MYB30 in the phloem accelerate flowering in Arabidopsis through the regulation of FLOWERING LOCUS T. PloS one 9(2):e89799
Long M, Rosenberg C, Gilbert W (1995) Intron phase correlations and the evolution of the intron/exon structure of genes. Proc Natl Acad Sci 92(26):12495–12499
Lynch M (2002) Intron evolution as a population-genetic process. Proc Natl Acad Sci 99(9):6118–6123
Mach M (2019) Camelina: a history of polyploidy, chromosome shattering, and recovery. Plant Cell 31(11):2550–2551
Maier A, Schrader A, Kokkelink L, Falke C, Welter B, Iniesto E, Rubio V, Uhrig JF, Hülskamp M, Hoecker U (2013) Light and the E3 ubiquitin ligase COP 1/SPA control the protein stability of the MYB transcription factors PAP 1 and PAP 2 involved in anthocyanin accumulation in Arabidopsis. Plant J 74(4):638–651
Makkena S, Lee E, Sack FD, Lamb RS (2012) The R2R3 MYB transcription factors FOUR LIPS and MYB88 regulate female reproductive development. J Exp Bot 63(15):5545–5558
Mandakova T, Pouch M, Brock JR, Al-Shehbaz IA, Lysak MA (2019) Origin and evolution of diploid and allopolyploid Camelina genomes were accompanied by chromosome shattering. Plant Cell 31(11):2596–2612
Marks D, Hopf T, Sander C (2012) Protein structure prediction from sequence variation. Nat Biotechnol 30:1072–1080
Marks DS, Colwell LJ, Sheridan R, Hopf TA, Pagnani A, Zecchina R, Sander C (2011) Protein 3D Structure Computed from Evolutionary Sequence Variation. PLoS One 6(12): e28766
Martín G, Márquez Y, Mantica F, Duque P, Irimia M (2021) Alternative splicing landscapes in Arabidopsis thaliana across tissues and stress conditions highlight major functional differences with animals. Genome Biol 22(1):1–26
Mayrose I, Lysak MA (2021) The evolution of chromosome numbers: mechanistic models and experimental approaches. Genome Biology and Evolution 13(2):evaa220
Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, Marshall WF (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318(5848):245–250
Millar AA, Gubler F (2005) The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17(3):705–721
Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carré IA, Coupland G (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2(5):629–641
Mohrmann L, Kal AJ, Verrijzer CP (2002) Characterization of the extended Myb-like DNA-binding domain of trithorax group protein Zeste. J Biol Chem 277(49):47385–47392
Mozgova I, Schrumpfová PP, Hofr C, Fajkus J (2008) Functional characterization of domains in AtTRB1, a putative telomere-binding protein in Arabidopsis thaliana. Phytochemistry 69(9):1814–1819
Mu RL, Cao YR, Liu YF, Lei G, Zou HF, Liao Y, Chen SY (2009) An R2R3-type transcription factor gene AtMYB59 regulates root growth and cell cycle progression in Arabidopsis. Cell Res 19(11):1291–1304
Muzyka A, Bazna A, Semilekto Y, Yemelyanov A (2012) Prism 6 for Windows. 6.GraphPad Software. Inc., San Diego, CA, USA
Nguyen HD, Yoshihama M, Kenmochi N (2006) Phase distribution of spliceosomal introns: implications for intron origin. BMC Evol Biol 6:69. https://doi.org/10.1186/1471-2148-6-69
Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, Vicedomini R (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497(7451):579–584
Ogata K, Hojo H, Aimoto S, Nakai T, Nakamura H, Sarai A, Ishii S, Nishimura Y (1992) Solution structure of a DNA-binding unit of Myb: a helix-turn-helix-related motif with conserved tryptophans forming a hydrophobic core. Proc Natl Acad Sci USA 89(14):6428–6432
Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y (1994) Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79(4):639–648
Oshima Y, Mitsuda N (2013) The MIXTA-like transcription factor MYB16 is a major regulator of cuticle formation in vegetative organs. Plant Signaling & Behavior 8(11):e26826
Palmer CM, Hindt MN, Schmidt H, Clemens S, Guerinot ML (2013) MYB10 and MYB72 are required for growth under iron-limiting conditions. PLoS Genetics 9(11):e1003953
Park MJ, Kwon YJ, Gil KE (2016) LATE ELONGATED HYPOCOTYL regulates photoperiodic flowering via the circadian clock in Arabidopsis. BMC Plant Biol 16:114
Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE (2021) UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30(1):70–82
Prouse MB, Campbell MM (2012) The interaction between MYB proteins and their target DNA binding sites. Biochim et Biophys Acta (BBA)-Gene Regulatory Mechanisms 1819(1):67–77
Punwani JA, Rabiger DS, Lloyd A, Drews GN (2008) The MYB98 subcircuit of the synergid gene regulatory network includes genes directly and indirectly regulated by MYB98. Plant J 55(3):406–414
Rabiger DS, Drews GN (2013) MYB64 and MYB119 are required for cellularization and differentiation during female gametogenesis in Arabidopsis thaliana. PLoS Genetics 9(9):e1003783
Rambaut A, Drummond AJ (2012) FigTree v1.4.4. Institute of Evolutionary Biology, University of Edinburgh, UK (http://tree.bio.ed.ac.uk/software/figtree/)
Ren R, Wang H, Guo C, Zhang N, Zeng L, Chen Y, Ma H, Qi J (2018) Widespread whole genome duplications contribute to genome complexity and species diversity in angiosperms. Mol Plant 11:414–428
Rensing SA, Goffinet B, Meyberg R, Wu SZ, Bezanilla M (2020) The moss Physcomitrium (Physcomitrella) patens: a model organism for non-seed plants. Plant Cell 32(5):1361–1376
Rosinski JA, Atchley WR (1998) Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. Jour Mol Evol 46(1):74–83
Roy SW, Penny D (2007) Patterns of intron loss and gain in plants: intron loss-dominated evolution and genome-wide comparison of O. sativa and A. thaliana. Mol Biol Evol 24(1):171–81
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34(12):3299–3302
Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15(16):2122–2133
Sadowski MI, Jones DT (2009) The sequence-structure relationship and protein function prediction. Curr Opin Struct Biol 19(3):357–362
Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, Minx P (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326(5956):1112–1115
Schranz M, Lysak M, Mitchell-Olds T (2006) The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci 11:532–542
Seeger M, Kraft R, Ferrell K, Bech-Otschir D, Dumdey R, Schade R, Gordon C, Naumann M, Dubiel W (1998) A novel protein complex involved in signal transduction possessing similarities to 26S proteasome subunits. FASEB J 12(6):469–478
Slotte T, Hazzouri KM, Ågren JA, Koenig D, Maumus F, Guo YL, Steige K, Platts AE, Escobar JS, Newman LK, Wang W (2013) The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat Genet 45(7):831–835
Solovyev V, Kosarev P, Seledsov I, Vorobyev D (2006) Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol 7(1):10–12
Stevenson CE, Burton N, Costa MM, Nath U, Dixon RA, Coen ES, Lawson DM (2006) Crystal structure of the MYB domain of the RAD transcription factor from Antirrhinum majus. Proteins: Structure, Function, and Bioinformatics 65(4):1041–5
Stracke R, Jahns O, Keck M, Tohge T, Niehaus K, Fernie AR, Weisshaar B (2010) Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12-, and MYB111-independent flavonol glycoside accumulation. New Phytol 188(4):985–1000
Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4(5):447–456
Sverdlov AV, Csuros M, Rogozin IB, Koonin EV (2007) A glimpse of a putative pre-intron phase of eukaryotic evolution. Trends Genet 23(3):105–108
Tanikawa J, Yasukawa T, Enari M, Ogata K, Nishimura Y, Ishii S, Sarai A (1993) Recognition of specific DNA sequences by the c-myb protooncogene product: role of three repeat units in the DNA-binding domain. Proc Natl Acad Sci 90(20):9320–9324
Thieme CJ, Rojas-Triana M, Stecyk E, Schudoma C, Zhang W, Yang L, Miñambres M, Walther D, Schulze WX, Paz-Ares J, Scheible WR, Kragler F (2015) Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat Plants 1(4):15025
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93(1):77–78
Wang B, Luo Q, Li Y, Yin L, Zhou N, Li X, Gan J, Dong A (2020a) Structural insights into target DNA recognition by R2R3-MYB transcription factors. Nucleic Acids Res 48(1):460–471
Wang S, Chen JG (2014) Regulation of cell fate determination by single-repeat R3 MYB transcription factors in Arabidopsis. Front Plant Sci 5:133
Wang X, Niu QW, Teng C, Li C, Mu J, Chua NH, Zuo J (2009) Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in Arabidopsis. Cell Res 19(2):224–235
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F, Huang S (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43(10):1035–1039
Wang XC, Wu J, Guan ML, Zhao CH, Geng P, Zhao Q (2020b) Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J 101(3):637–652
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TA, Rempfer C, Bordoli L, Lepore R (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46(W1):W296-303
Wendel JF, Jackson SA, Meyers BC, Wing RA (2016) Evolution of plant genome architecture. Genome Biol 17:37
Wikstrom MK (1977) Proton pump coupled to cytochrome c oxidase in mitochondria. Nature 266(5599):271–273
Williams KL (2019) Gene Mapping. In S. Ranganathan, M. Gribskov, K. Nakai, C. Schönbach, & B. Gaeta (Eds.), Encyclopedia of bioinformatics and computational biology (Vol. 2, pp. 242–250). Academic Press. https://doi.org/10.1016/B978-0-12-809633-8.20233-1
Wu P, Peng M, Li Z, Yuan N, Hu Q, Foster CE, Saski C, Wu G, Sun D, Luo H (2019) DRMY1, a Myb-like protein, regulates cell expansion and seed production in Arabidopsis thaliana. Plant Cell Physiol 60(2):285–302
Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PloS One 2(8):e718
Xu R, Wang Y, Zheng H, Lu W, Wu C, Huang J, Yan K, Yang G, Zheng C (2015) Salt-induced transcription factor MYB74 is regulated by the RNA-directed DNA methylation pathway in Arabidopsis. J Exp Bot 66(19):5997–6008
Yang C, Xu Z, Song J, Conner K, Vizcay BG, Wilson ZA (2007) Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19(2):534–548
Yang R, Jarvis DJ, Chen H, Beilstein M, Grimwood J, Jenkins J, Shu S, Prochnik S, Xin M, Ma C, Schmutz J (2013) The reference genome of the halophytic plant Eutrema salsugineum. Front Plant Sci 4:46
Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, Zhiqiang L, Yunfei Z, Xiaoxiao W, Xiaoming Q, Yunping S (2006) The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 60(1):107–124
Yao H, Guo L, Fu Y, Borsuk LA, Wen TJ, Skibbe DS, Cui X, Scheffler BE, Cao J, Emrich SJ, Ashlock DA (2005) Evaluation of five ab initio gene prediction programs for the discovery of maize genes. Plant Mol Biol 57(3):445–460
Yatusevich R (2008) Analysis of the MYB28, MYB29 and MYB76 transcription factors involved in the biosynthesis of aliphatic glucosinolates in Arabidopsis thaliana (Doctoral dissertation, Universität zu Köln)
Yu A, Li F, Xu W, Wang Z, Sun C, Han B, Wang Y, Wang B, Cheng X, Liu A (2019) Application of a high-resolution genetic map for chromosome-scale genome assembly and fine QTLs mapping of seed size and weight traits in castor bean. Sci Rep 9(1):1–1
Zaware N, Zhou MM (2019) Bromodomain biology and drug discovery. Nat Struct Mol Biol 26(10):870–879
Zhang P, Wang R, Ju Q, Li W, Tran LSP, Xu J (2019) The R2R3-MYB transcription factor MYB49 regulates cadmium accumulation. Plant Physiol 180(1):529–542
Zhou J, Lee C, Zhong R, Ye ZH (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21(1):248–266
Zhu Y, Rong L, Luo Q, Wang B, Zhou N, Yang Y, Dong A (2017) The histone chaperone NRP1 interacts with WEREWOLF to activate GLABRA2 in Arabidopsis root hair development. Plant Cell 29(2):260–276
Funding
The award”” of JRF/SRF from UGC to Mukund Lal, from CSIR to Nishu Chahar, and Shobha Yadav, and SRF from DBT (BT/PR18086/BPA/118/186/2016) to Ekta Bhardwaj is gratefully acknowledged. Sandip Das is financially supported by the Institute of Eminence (IoE), University of Delhi (IoE/FRP/LS/2020/27).
Author information
Authors and Affiliations
Contributions
ML and SD designed the study. ML performed all analysis with help from EB and NC (identification of homologs, analysis of genic structure and features, phylogeny and protein attributes) and SY (identification of tandem genes and synteny). ML and SD analysed all data and wrote the MS. All authors have read and agree with the findings.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10142_2022_836_MOESM1_ESM.pdf
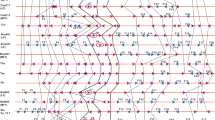
Supplementary file1 (PDF 1328 kb) Phylogram of 2R-MYBs to identify orthologs and paralogs from A. thaliana-A. lyrata [A]; A. thaliana-C. rubella [B]; A. thaliana-C. sativa [C]; A. thaliana-E. salsugineum [D]; and A. thaliana-B. rapa [E].
10142_2022_836_MOESM2_ESM.pdf
Supplementary file2 (PDF 4914 kb) Mapping of distribution and organisation of 1R-MYBs on pseudochromosomes of M. polymorpha, S. moellendorffii, P. abies, A. trichopoda, P. patens, Z. mays, and A. thaliana [A-G]; and 2R-MYB genes on pseudochromosomes of A. thaliana, A. lyrata, C. rubella, C. sativa, and E. salsugineum [H-L].
10142_2022_836_MOESM3_ESM.pdf
Supplementary file3 (PDF 512 kb) Physico-chemical properties of 1R-MYBs in eight plant species and 2R-MYBs in six Brassicaceae species. Bar graphs represent molecular weight of 1R-MYB [A] genes are greater than 2R-MYBs [D]; pI values range from 4 to 10 in both 1R- and 2R-MYBs [B and E]; and GRAVY values for most 1R-MYB and 2R-MYB genes range from -1.25 to -0.25 and -1.0 to -0.5, respectively [C and F].
10142_2022_836_MOESM4_ESM.pdf
Supplementary file4 (PDF 428 kb) Length and position of MYB domain in 1R-MYBs from eight plant species and 2R-MYBs from six Brassicaceae species. Bar graphs show that the length of most 1R-MYB and 2R-MYB genes range from 40 aa to 60 aa and 95 aa to 100 aa, respectively [A and C]; besides the position of the MYB domain in the most 1R- and 2R-MYBs is towards N-terminal region, whereas several 1R-MYB genes have MYB-domain positioned at the centre or towards the C-terminal [B and D].
10142_2022_836_MOESM5_ESM.pdf
Supplementary file5 (PDF 1091 kb) Exons-intron structure, intron phases, and conserved MYB domain using genomic sequences, CDS, and coordinates of MYB domain repeat sequences of 1R-MYBs from eight plant species i.e., C. reinhardtii [A], M. polymorpha [B], P. patens [C], S. moellendorffii [D], P. abies [E], A. trichopoda [F], Z. mays [G], and A. thaliana [H]; and 2R-MYBs from 6 Brassicaceae species i.e., A. thaliana [I], A. lyrata [J], C. rubella [K], C. sativa [L], E. salsugineum [M], and B. rapa [N].
10142_2022_836_MOESM6_ESM.pdf
Supplementary file6 (PDF 349 kb) Phylogenetic reconstruction of MYB repeat sequences (R1, R2 and R3-type) using Neighbor-Joining method with JTT matrix-based model
10142_2022_836_MOESM7_ESM.pdf
Supplementary file7 (PDF 203 kb) Clade wise representation of the conserved MYB domain and C-terminal motifs of 2R-MYBs from A. thaliana
10142_2022_836_MOESM8_ESM.pdf
Supplementary file8 (PDF 1092 kb) Phylogenetic reconstruction of 2R-MYB genes from A. thaliana using Neighbor-Joining method with JTT matrix-based model displays extensive paralogy; clustering based on sequence similarity and similar gene features; and homologs in a given cluster also share functional similarity.
10142_2022_836_MOESM9_ESM.pdf
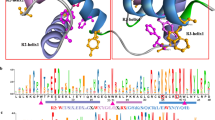
Supplementary file9 (PDF 2128 kb) Clade-wise alignment of the MYB domain with R2 and R3 repeats and web-logo showing conserved and regularly spaced Tryptophan residues (W) in A. thaliana 2R-MYB genes.
10142_2022_836_MOESM10_ESM.pdf
Supplementary file10 (PDF 957 kb) Heat-maps showing differential expression of thirteen 2R-MYB genes from A. thaliana under three abiotic stress conditions-cold [A], salt [B], and drought [C]. Blue and yellow colour represent the least and the highest Z-score, respectively. The downregulation and upregulation of the gene expression are represented as red and green arrows, respectively.
10142_2022_836_MOESM11_ESM.pdf
Supplementary file11 (PDF 510 kb) Comparative genomics of tandemly organised 2R-MYBs -AtMYB23-AtMYB24-AtMYB115, and AtMYB104-AtMYB81 [A], employing micro-synteny analysis using ca. 100 kb genomic segments across different genomes of Brassicaceae reveals gene losses, duplication and rearrangements [B-C]. Genes unique to a particular genomic segment is marked by black arrows. Larger and coloured arrows represent tandem 2R-MYB genes under investigation. Smaller coloured arrows represent genes that are conserved across at least two genomic segments. Blue rectangle represents duplicated genes whereas red circle represents the most abundant gene.
10142_2022_836_MOESM13_ESM.pdf
Supplementary file13 (PDF 1599 kb) Distribution of 2R-MYBs across the 24-genomic blocks (A-X) across Brassicaceae. Homologous blocks are boxed with similar colours.
10142_2022_836_MOESM14_ESM.xlsx
Supplementary file14 (XLSX 277 kb) Identification of all MYB genes (1R-, 2R-, 3R- and atypical-MYBs) and their genomic features -number of genes; length of gene, CDS and protein; gene/CDS ratio; intron number and phases; molecular weight; pI values; GRAVY values; MYB domain length and its position across 16 taxa representing protists, fungi, animals and plants.
10142_2022_836_MOESM15_ESM.xlsx
Supplementary file15 (XLSX 618 kb) Identification of 2R-MYB genes and their genomic features such as number of genes; length of gene, CDS and protein; gene/CDS ratio; intron number and phases; molecular weight; pI values; GRAVY values; MYB domain length and its position across 6 taxa representing family Brassicaceae.
10142_2022_836_MOESM16_ESM.docx
Supplementary file16 (DOCX 19 kb) Summary of 3D-structure models of clade-wise consensus MYB domain sequences of 1R-MYBs [A] and 2R-MYBs [B] from A. thaliana.
10142_2022_836_MOESM17_ESM.xlsx
Supplementary file17 (XLSX 37 kb) Compiled information about the paralogy, track-files, and label-files of 1R-MYBs and 2R-MYBs from A. thaliana.
10142_2022_836_MOESM18_ESM.xlsx
Supplementary file18 (XLSX 1098 kb) Analysis of the frequency distributions of synonymous substitutions (Ks) and its ratio with non-synonymous substitutions (Ka/Ks; ω) for 1R-MYB paralogs in A. thaliana; 2R-MYB paralogs in A. thaliana and their orthologs in A. lyrata, C. rubella, C. sativa, E. salsugineum, and B. rapa
10142_2022_836_MOESM19_ESM.xlsx
Supplementary file19 (XLSX 34 kb) Normalised microarray expression data for 13 selected A. thaliana 2R-MYB genes belonging to clade 2 (AtMYB49, AtMYB74 and AtMYB102), clade 6 (AtMYB10, AtMYB13, AtMYB14, AtMYB15, AtMYB60 and AtMYB72) and clade 14 (AtMYB44, AtMYB70, AtMYB73 and AtMYB77) that act as regulators under cold, salt and drought stress conditions.
10142_2022_836_MOESM20_ESM.xlsx
Supplementary file20 (XLSX 58 kb) Gene list for Micro-synteny analysis of the 2R-MYB gene pair AtMYB113-AtMYB114-AtMYB90.
10142_2022_836_MOESM23_ESM.xlsx
Supplementary file23 (XLSX 40 kb) Gene list for micro-synteny analysis of the 2R-MYB gene pair AtMYB23-AtMYB24-AtMYB115.
Rights and permissions
About this article
Cite this article
Lal, M., Bhardwaj, E., Chahar, N. et al. Comprehensive analysis of 1R- and 2R-MYBs reveals novel genic and protein features, complex organisation, selective expansion and insights into evolutionary tendencies. Funct Integr Genomics 22, 371–405 (2022). https://doi.org/10.1007/s10142-022-00836-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-022-00836-w