Abstract
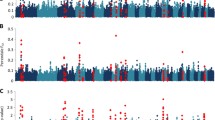
Gastrointestinal (GI) nematode infections are a worldwide threat to human health and animal production. In this study, we performed a genome-wide association study between copy number variations (CNVs) and resistance to GI nematodes in an Angus cattle population. Using a linear regression analysis, we identified one deletion CNV which reaches genome-wide significance after Bonferroni correction. With multiple mapped human olfactory receptor genes but no annotated bovine genes in the region, this significantly associated CNV displays high population frequencies (58.26 %) with a length of 104.8 kb on chr7. We further investigated the linkage disequilibrium (LD) relationships between this CNV and its nearby single nucleotide polymorphisms (SNPs) and genes. The underlining haplotype blocks contain immune-related genes such as ZNF496 and NLRP3. As this CNV co-segregates with linked SNPs and associated genes, we suspect that it could contribute to the detected variations in gene expression and thus differences in host parasite resistance.


Similar content being viewed by others
Abbreviations
- CNV:
-
Copy number variation
- CNVR:
-
CNV region
- GI:
-
Gastrointestinal
- SNP:
-
Single nucleotide polymorphism
References
Araujo RN, Padilha T, Zarlenga D, Sonstegard T, Connor EE, Van Tassel C, Lima WS, Nascimento E, Gasbarre LC (2009) Use of a candidate gene array to delineate gene expression patterns in cattle selected for resistance or susceptibility to intestinal nematodes. Vet Parasitol 162:106–115
Bae JS, Cheong HS, Kim LH, NamGung S, Park TJ, Chun JY, Kim JY, Pasaje CF, Lee JS, Shin HD (2010) Identification of copy number variations and common deletion polymorphisms in cattle. BMC Genomics 11:232
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265
Bickhart DM, Hou Y, Schroeder SG, Alkan C, Cardone MF, Matukumalli LK, Song J, Schnabel RD, Ventura M, Taylor JF, Garcia JF, Van Tassell CP, Sonstegard TS, Eichler EE, Liu GE (2012) Copy number variation of individual cattle genomes using next-generation sequencing. Genome Res 22:778–790
Cicconardi F, Chillemi G, Tramontano A, Marchitelli C, Valentini A, Ajmone-Marsan P, Nardone A (2013) Massive screening of copy number population-scale variation in Bos taurus genome. BMC Genomics 14:124
Coppieters W, Mes TH, Druet T, Farnir F, Tamma N, Schrooten C, Cornelissen AW, Georges M, Ploeger HW (2009) Mapping QTL influencing gastrointestinal nematode burden in Dutch Holstein-Friesian dairy cattle. BMC Genomics 10:96
Crawford AM, Paterson KA, Dodds KG, Diez TC, Williamson PA, Roberts TM, Bisset SA, Beattie AE, Greer GJ, Green RS, Wheeler R, Shaw RJ, Knowler K, McEwan JC (2006) Discovery of quantitative trait loci for resistance to parasitic nematode infection in sheep: I. Analysis of outcross pedigrees. BMC Genomics 7:178
Diez-Tascon C, Keane OM, Wilson T, Zadissa A, Hyndman DL, Baird DB, McEwan JC, Crawford AM (2005) Microarray analysis of selection lines from outbred populations to identify genes involved with nematode parasite resistance in sheep. Physiol Genomics 21:59–69
Fadista J, Thomsen B, Holm LE, Bendixen C (2010) Copy number variation in the bovine genome. BMC Genomics 11:284
Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229
Gasbarre LC, Leighton EA, Sonstegard T (2001) Role of the bovine immune system and genome in resistance to gastrointestinal nematodes. Vet Parasitol 98:51–64
Glick G, Shirak A, Seroussi E, Zeron Y, Ezra E, Weller JI, Ron M (2011) Fine mapping of a QTL for fertility on BTA7 and its association with a CNV in the Israeli Holsteins. G3 (Bethesda) 1:65–74
Harhay GP, Smith TP, Alexander LJ, Haudenschild CD, Keele JW, Matukumalli LK, Schroeder SG, Van Tassell CP, Gresham CR, Bridges SM, Burgess SC, Sonstegard TS (2010) An atlas of bovine gene expression reveals novel distinctive tissue characteristics and evidence for improving genome annotation. Genome Biol 11:R102
Henrichsen CN, Vinckenbosch N, Zollner S, Chaignat E, Pradervand S, Schutz F, Ruedi M, Kaessmann H, Reymond A (2009) Segmental copy number variation shapes tissue transcriptomes. Nat Genet 41:424–429
Hou Y, Liu GE, Bickhart DM, Cardone MF, Wang K, Kim ES, Matukumalli LK, Ventura M, Song J, VanRaden PM, Sonstegard TS, Van Tassell CP (2011a) Genomic characteristics of cattle copy number variations. BMC Genomics 12:127
Hou Y, Liu GE, Bickhart DM, Matukumalli LK, Li C, Song J, Gasberre LC, Van Tassell CP, Sonstegard TS (2011b) Genomic regions showing copy number variations associate with resistance or susceptibility to gastrointestinal nematodes in Angus cattle. Funct Integr Genomics 12:81–92
Hou Y, Bickhart DM, Hvinden ML, Li C, Song J, Boichard DA, Fritz S, Eggen A, Denise S, Wiggans GR, Sonstegard TS, Van Tassell CP, Liu GE (2012) Fine mapping of copy number variations on two cattle genome assemblies using high density SNP array. BMC Genomics 13:376
Ingham A, Reverter A, Windon R, Hunt P, Menzies M (2008) Gastrointestinal nematode challenge induces some conserved gene expression changes in the gut mucosa of genetically resistant sheep. Int J Parasitol 38:431–442
Jackson JA, Turner JD, Kamal M, Wright V, Bickle Q, Else KJ, Ramsan M, Bradley JE (2006) Gastrointestinal nematode infection is associated with variation in innate immune responsiveness. Microbes Infect 8:487–492
Kadri NK, Koks PD, Meuwissen TH (2012) Prediction of a deletion copy number variant by a dense SNP panel. Genet Sel Evol 44:7
Keane OM, Zadissa A, Wilson T, Hyndman DL, Greer GJ, Baird DB, McCulloch AF, Crawford AM, McEwan JC (2006) Gene expression profiling of naive sheep genetically resistant and susceptible to gastrointestinal nematodes. BMC Genomics 7:42
Kim E, Sonstegard TS, Silva MVGB, Gasbarre LC, Van Tassell CP (2013) Identification of QTL affecting gastrointestinal nematodes resistance in an experimental Angus population. Animal Genetics Accepted
Li RW, Gasbarre LC (2009) A temporal shift in regulatory networks and pathways in the bovine small intestine during Cooperia oncophora infection. Int J Parasitol 39:813–824
Li RW, Sonstegard TS, Van Tassell CP, Gasbarre LC (2007) Local inflammation as a possible mechanism of resistance to gastrointestinal nematodes in Angus heifers. Vet Parasitol 145:100–107
Liu GE, Hou Y, Zhu B, Cardone MF, Jiang L, Cellamare A, Mitra A, Alexander LJ, Coutinho LL, Dell’aquila ME, Gasbarre LC, Lacalandra G, Li RW, Matukumalli LK, Nonneman D, Regitano LC, Smith TP, Song J, Sonstegard TS, Van Tassell CP, Ventura M, Eichler EE, McDaneld TG, Keele JW (2010) Analysis of copy number variations among diverse cattle breeds. Genome Res 20:693–703
Liu GE, Brown T, Hebert DA, Cardone MF, Hou YL, Choudhary RK, Shaffer J, Amazu C, Connor EE, Ventura M, Gasbarre LC (2011) Initial analysis of copy number variations in cattle selected for resistance or susceptibility to intestinal nematodes. Mamm Genome 22:111–121
Menzies M, Reverter A, Andronicos N, Hunt P, Windon R, Ingham A (2010) Nematode challenge induces differential expression of oxidant, antioxidant and mucous genes down the longitudinal axis of the sheep gut. Parasite Immunol 32:36–46
Scherer SW, Lee C, Birney E, Altshuler DM, Eichler EE, Carter NP, Hurles ME, Feuk L (2007) Challenges and standards in integrating surveys of structural variation. Nat Genet 39:S7–S15
Seroussi E, Glick G, Shirak A, Yakobson E, Weller JI, Ezra E, Zeron Y (2010) Analysis of copy loss and gain variations in Holstein cattle autosomes using BeadChip SNPs. BMC Genomics 11:673
Sonstegard TS, Gasbarre LC (2001) Genomic tools to improve parasite resistance. Vet Parasitol 101:387–403
Stepek G, Buttle DJ, Duce IR, Behnke JM (2006) Human gastrointestinal nematode infections: are new control methods required? Int J Exp Pathol 87:325–341
Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, Tyler-Smith C, Carter N, Scherer SW, Tavare S, Deloukas P, Hurles ME, Dermitzakis ET (2007) Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315:848–853
Zhang F, Gu W, Hurles ME, Lupski JR (2009) Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet 10:451–481
Acknowledgments
We thank Reuben Anderson and Alexandre Dimitriv for their technical assistance. This work was supported in part by AFRI grant no. 2011-67015-30183 from USDA NIFA. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. The USDA is an equal opportunity provider and employer.
Conflict of interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xu, L., Hou, Y., Bickhart, D.M. et al. A genome-wide survey reveals a deletion polymorphism associated with resistance to gastrointestinal nematodes in Angus cattle. Funct Integr Genomics 14, 333–339 (2014). https://doi.org/10.1007/s10142-014-0371-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-014-0371-6