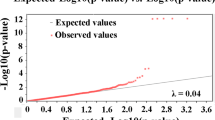
Abstract
The large yellow croaker is one of the largest marine economic fish in China with the farming yield about 30 tons per year. The genetic selection for growth and disease resistance has been performed in recent years. The identification of trait-associated molecular makers, causative variants, and causative genes is helpful for genetic selection in large yellow croaker. It has been discovered that most of polygenic traits are controlled with multiple genes via regulatory variant, and GWAS-identified loci are enriched in the regulatory variant. Cis-acting eQTL is a widespread regulatory variant that controls the expression of nearby gene. We herein take advantage of RNA-seq and whole genome sequencing technique to identify genome-wide eQTLs in liver tissue for large yellow croaker; a forward selection routine is applied for identification of multiple eQTLs. To fine map causative mutation for each eQTL, a credible set is built to confine causative variants. Totally, 2427 eQTLs have been identified, 69.7% (1,691/2,427) of them are primary eQTL signals, and the remaining are secondary signals, many functional important target genes have been discovered. We highlight several functional pivotal genes including SMC3, TUSC3, TITIN, MCPH1, and MDHC, in which the expression of MCPH1 is regulated by two eQTLs; the distance of these eQTLs from target genes is symmetrically distributed, 25.5% of them are within 1 Mb region from target genes, whereas 74.5% of them are between 1 and 2 Mb regions; most of the identified eQTL has been well resolved, and 19.3 (469/2427) of eQTL have the size of credible set (the number of variants in credible set) less than 50.



Similar content being viewed by others
References
Aguet F et al (2017) Genetic effects on gene expression across human tissues. Nature 550:204–213
Ardlie KG et al (2015) The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348:648–660
Bolger A, Lohse M, Usadel B (2014) Trimmomatic: flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Davies RW, Flint J, Myers S, Mott R (2016) Rapid genotype imputation from sequence without reference panels. Nat Genet 48:965–969
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C et al (2012) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21
Dong LS, Fang M, Wang ZY (2017) Prediction of genomic breeding values using new computing strategies for the implementation of MixP. Sci Rep 7:17200
Eckels EC, Tapia-Rojo R, Rivas-Pardo JA, Fernández JM (2018) The work of titin protein folding as a major driver in muscle contraction. Annu Rev Physiol 80:327–351
eGTEx Project (2017) Enhancing GTEx by bridging the gaps between genotype, gene expression, and disease. Nat Genet 49:1664–1670
Fang M, Georges M (2016) BayesFM: a software program to fine-map multiple causative variants in GWAS identified risk loci. biorxiv. https://doi.org/10.1101/067801
Frank WA, Kruglyak L (2015) The role of regulatory variation in complex traits and disease. Nat Rev Genet 16:197–212
Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, Eyler AE, Denny JC, Nicolae DL, Cox NJ, Im HK (2015) A gene-based association method for mapping traits using reference transcriptome data. Nat Genet 47:1091–1098
Gao YX, Dong LS, Xu SB, Xiao SJ, Fang M, Wang ZY (2018) Genome-wide association study using single marker analysis and Bayesian methods for the gonadosomatic index in the large yellow croaker. Aquaculture 486:26–30
Ghiselli G (2006) SMC3 knockdown triggers genomic instability and p53-dependent apoptosis in human and zebrafish cells. Mol Cancer 5:52
Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, Plagnol V (2014) Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLos Genet 10:e1004383
GTEx Consortium, Lead analysts ,Laboratory, Data Analysis & Coordinating Center (LDACC) et al . (2017) Genetic effects on gene expression across human tissues. Nature 550:204–213
Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BWJH, Jansen R, de Geus EJC, Boomsma DI, Wright FA, Sullivan PF, Nikkola E, Alvarez M, Civelek M, Lusis AJ, Lehtimäki T, Raitoharju E, Kähönen M, Seppälä I, Raitakari OT, Kuusisto J, Laakso M, Price AL, Pajukanta P, Pasaniuc B (2016) Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet 48:245–252
Hoffman JD, Graff RE, Emami NC, Tai CG, Passarelli MN, Hu D, Huntsman S, Hadley D, Leong L, Majumdar A, Zaitlen N, Ziv E, Witte JS (2017) Cis-eQTL-based trans-ethnic meta-analysis reveals novel genes associated with breast cancer risk. PLos Genet 13:e1006690
Huang H, Fang M, Jostins L, Mirkov MU, Boucher G et al (2017) Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 547:173–178
Jo YH, Kim HO, Lee J, Lee SS, Cho CH, Kang IS, Choe WJ, Baik HH, Yoon KS (2013) MCPH1 protein expression and polymorphisms are associated with risk of breast cancer. Gene 517:184–190
Jo YS, Kim SS, Kim MS, Yoo NJ, Lee SH (2017) Candidate tumor suppressor gene MCPH1 is mutated in colorectal and gastric cancers. Int J Colorectal Dis 32:161–162
Kuroha T, Nagai K, Kurokawa Y, Nagamura Y, Kusan, M, Yasu, H, Fukushima A (2017) eQTLs regulating transcript variations associated with rapid internode elongation in deepwater rice. Front Plant Scie 8:1753
Li X, Kim Y, Tsang EK, Davis JR, Damani FN, Chiang C, Montgomery SB (2017) The impact of rare variation on gene expression across tissues. Nature 550:239–243
Linke WA (2018) Titin gene and protein functions in passive and active muscle. Annu Rev Physiol 80:389–411
Liu Y, Liu XL, Zheng ZW, Ma TT, Liu Y (2020) Genome-wide analysis of expression QTL (eQTL) and allele-specific expression (ASE) in pig muscle identifies candidate genes for meat quality traits. Genet Sel Evol 52:59
Mancuso N, Shi H, Goddard P, Kichaev G, Gusev A, Pasaniuc B (2017) Integrating gene expression with summary association statistics to identify genes associated with 30 complex traits. Am J Hum Genet 100:473–487
Mohammadi P, Castel SE, Brown AA, Lappalainen T (2017) Quantifying the regulatory effect size of cis-acting genetic variation using allelic fold change. Genome Res 27:1872–1884
Momozawa Y, Dmitrieva J, Théâtre E, Deffontaine V, Rahmouni S, Charloteaux B, Crins F, Docampo E, Elansary M, Gori AS, Lecut C, Mariman R, Mni M, Oury C, Altukhov I, Alexeev D, Aulchenko Y, Amininejad L, Bouma G, Hoentjen F, Löwenberg M, Oldenburg M, Pierik MJ, Vander AE, de, Jong MC, van der Woude J, Visschedijk MC, The International IBDGeneticsConsortium, Lathrop M, Hugot JP, Weersma RK, Vos MD, Franchimont D, Vermeire S, Kubo M, Louis E, Georges M (2018) IBD risk loci are enriched in multigenic regulatory modules encompassing putative causative genes. Nat Commun 9:2427
Naseer MI, Rasool M, Abdulkareem AA, Bassiouni RI, Algahtani H, Chaudhary AG, Al-Qahtani MH (2018) Novel compound heterozygous mutations in MCPH1 gene causes primary microcephaly in Saudi family. Neurosciences 23:347–350
Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ (2010) Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLos Genet 6:e1000888
Oliva M, Muñoz-Aguirre M, Kim-Hellmuth S, Wucher V, Gewirtz ADH et al (2020) The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369:1318–1330
Peng Y, Cao J, Yao XY, Wang JX, Zhong MZ, Gan PP, Li JH (2017) TUSC3 induces autophagy in human non-small cell lung cancer cells through Wnt/β-catenin signaling. Oncotarget 8:52960–52974
Pils D, Horak P, Vanhara P, Anees M, Petz M, Alfanz A, Gugerell A, Wittinger M, Gleiss A, Auner V, Tong D, Zeillinger R, Braicu EI, Sehouli J, Krainer M (2013) Methylation status of TUSC3 is a prognostic factor in ovarian cancer. Cancer 119:946–954
Ponsuksili S, Murani E, Schwerin M, Schellander K, Wimmers K (2010) Identification of expression QTL (eQTL) of genes expressed in porcine M. longissimus dorsi and associated with meat quality traits. BMC Genomics 11 (572)
Ren YS, Deng RX, Cai R, Lu XS, Luo YJ, Wang ZY, Zhu YC, Yin MY, Ding YQ, Lin J (2020) TUSC3 induces drug resistance and cellular stemness via Hedgehog signaling pathway in colorectal cancer. Carcinogenesis, bgaa038. https://doi.org/10.1093/carcin/bgaa038
Rimmer A, Phan H, Mathieson I, Iqbal Z, Twigg SR, Wilkie AO et al (2014) Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet 46:912–918
Savarese M, Maggi L, Vihola A, Jonson PH, Tasca G, Ruggiero L, Bello L, Magri F, Giugliano T, Torella A, Evilä A, Di FG, Vanakker O, Gibertini S, Vercelli L, Ruggieri A, Antozzi C, Luque H, Janssens S, Pasanisi MB, Fiorillo C, Raimondi M, Ergoli M, Politano L, Bruno C, Rubegni A, Pane M, Santorelli FM, Minetti C, Angelini C, De BJ, Moggio M, Mongini T, Comi GP, Santoro L, Mercuri E, Pegoraro E, Mora M, Hackman P, Udd B, Nigro V (2018) Interpreting Genetic Variants in Titin in Patients With Muscle Disorders. JAMA Neurol 75:557–565
Wang Y, Guo W, Xu H, Zhu X, Yu T, Jiang Z, Jiang Q, Gang X (2018) An extensive study of the mechanism of prostate cancer metastasis. Neoplasma 65:253–261
Wang T, Glover B, Hadwiger G, Miller CA, di Martino O, Welch JS (2019) Smc3 is required for mouse embryonic and adult hematopoiesis. Exp Hematol 70:70–84
Yang F, Wang J, Pierce BL, Chen LS (2017) Identifying cis-mediators for trans-eQTLs across many human tissues using genomic mediation analysis. Genome Res 27:1859–1871
Yuan J, Yu XS, Wang AH, Li Y, Liu FJ, Wang Y, Sun SM, Bing XY, Liu YM, Du J (2018) Tumor suppressor candidate 3: a novel grading tool and predictor of clinical malignancy in human gliomas. Oncol Lett 15:5655–5661
Funding
The research was supported by National Natural Science Foundation of China Grant 31672399 and 31872560, Natural Science Foundation of 2020J01672, 2018J01450, National Marine Fisheries Industrial Technology System Post Scientist Project (CARS-47-G04), and Xiamen Science and Technology Project (2019SH400133).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jiang, D., Li, W., Wang, Z. et al. Genome-Wide Identification of Cis-acting Expression QTLs in Large Yellow Croaker. Mar Biotechnol 23, 225–232 (2021). https://doi.org/10.1007/s10126-020-10017-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-020-10017-0