Abstract
Background
Children in the intensive care unit (ICU) who suffer from severe basic diseases and low immunity are usually in critical condition. It is crucial to assist clinicians in selecting the appropriate empirical antibiotic therapies for clinical infection control.
Methods
We retrospectively analyzed data from 281 children with bloodstream infection (BSI). Comparisons of basic data, pathogenic information, and drug resistance of the main bacteria were conducted.
Results
We detected 328 strains, including Gram-positive bacteria (223, 68%), mainly coagulase-negative Staphylococci (CoNS); Gram-negative bacteria (91, 27.7%); and fungi (14, 4.3%). The results of the binary logistic regression analysis showed that the main basic disease was an independent risk factor for death. Compared with Escherichia coli, Klebsiella pneumoniae exhibited a higher proportion of extended-spectrum β-lactamases (ESBLs), and its resistance to some β-lactamides and quinolones antibiotics were lower. Twenty-seven isolates of multidrug-resistant (MDR) bacteria were detected, of which carbapenem-resistant Acinetobacter baumannii (CRAB) accounted for the highest proportion (13, 48.2%).
Conclusions
CoNS was the principal pathogen causing BSI in children in the ICU of children, and Escherichia coli was the most common Gram-negative pathogen. The main basic disease was an independent risk factor for death. It is necessary to continuously monitor patients with positive blood cultures, pay special attention to detected MDR bacteria, and strengthen the management of antibiotics and prevention and control of nosocomial infections.
Similar content being viewed by others
Introduction
Bloodstream infection (BSI) is a systemic infectious disease caused by pathogenic microorganisms entering the blood system and can manifest as bacteremia or even sepsis (Gouel-Cheron et al. 2022). Sepsis is a serious systemic inflammatory reaction and is one of the main causes of death in children (Zhang et al. 2022). Children in the intensive care unit (ICU), often in critical condition with severe basic diseases and low immunity, are prone to infections (Yan et al. 2021). According to statistics, the hospital infection of children in the ICU is approximately 2–5 times that of children in general wards (Bassetti et al. 2015; Bammigatti et al. 2017). Once a child suffers from BSI, it not only aggravates the illness and elevates pain but also leads to extended hospital stays, significantly inflating medical expenses. This situation seriously threatens the life of the child and amplifies the financial burden on the family (Tran et al. 2017; Zhu et al. 2019). Recently, the incidence and mortality rates of BSI have remained high. Studies have shown that BSI is the most common hospital-acquired infection in the ICU, with a mortality of 18.6–52.3% (Shime et al. 2012; Marsillio et al. 2015; Schwab et al. 2018; Markwart et al. 2020). Therefore, early and appropriate antibiotic therapy can improve the prognosis of children with sepsis admitted to the ICU. Blood culture is considered the most effective laboratory method for diagnosing BSI. In clinical practice, pathogens are identified through blood culture results, and rational drug use is based on drug sensitivity results. However, the low positive rate of blood cultures and delayed reporting of positive drug sensitivity results pose a challenge. Therefore, it is difficult to swiftly and accurately guide the selection of antibiotics. Hence, it is crucial for clinicians to accurately evaluate the condition of children with BSI and rapidly understand pathogen characteristics and drug resistance. This allows for the empirical selection of antibiotics and enhancing the efficacy of infection treatments. Studies on BSI in ICU have been conducted. However, infection control and antimicrobial management policies differ among countries, regions, and hospitals, leading to distinct clinical characteristics among BSI pathogens (Timsit et al. 2020; Xie et al. 2020). The data survey results showed no reports on the characteristics and drug resistance of pathogenic bacteria of BSI in ICU children in the Suzhou area over the last 6 years. Therefore, the purpose of this study was to retrospectively analyze the distribution of pathogenic bacteria, risk factors for death, and drug resistance in children with BSI admitted to the ICU of the Children’s Hospital Affiliated with Suzhou University from January 2016 to December 2021. The aim was to guide clinicians in selecting appropriate empirical treatment schemes by observing the clinical symptoms of children and combining research data before receiving feedback from the laboratory to reduce the problem of antibiotic abuse and the production of multidrug-resistant (MDR) bacteria.
Materials and methods
Study site
This study was conducted at the Children’s Hospital of Soochow University, a medical center in East China and the only tertiary children’s hospital in Jiangsu Province. The Children’s Hospital of Soochow University has 1500 beds and serves > 70,000 inpatients and > 2 million outpatients annually. This study was approved by the Ethics Committee of the Children’s Hospital of Soochow University (No. 2021CS158).
General information
A total of 281 patients with BSI who were admitted to the ICU of the Children’s Hospital of Soochow University between January 2016 and December 2021 were selected for the study. According to the number of pathogenic bacteria isolated from blood culture samples, children with only one type of pathogenic bacteria in the blood culture samples were included in the single infection group (243 cases), while children with ≥ 2 pathogens cultured simultaneously or ≥ 2 pathogens isolated several times in a row were included in the mixed infection group (38 cases).
Diagnostic criteria for BSI
According to the national diagnostic standard for hospital infection (Diagnostic criteria for nosocomial infection (Trial), 2003), (Ministry of Health of the People’s Republic of China 2003) and the latest definition and diagnostic standard for hospital infection of CDC/NHSN in the USA (CDC/NHSN, 2017), if a patient had isolated pathogenic bacteria from blood samples during hospitalization and exhibited any of the following symptoms or signs: (1) temperature > 38 ℃ or < 36 ℃, accompanied by chills; (2) invasive portals or migratory lesions of pathogens; (3) obvious symptoms of systemic infection and poisoning, but no clear infection focus; and (4) systolic blood pressure lower than 90 mmHg or more than 40 mmHg lower than the original systolic blood pressure. The exclusion criteria were as follows: (1) incomplete case data, (2) elimination of contaminated strains, and (3) repeated detection of the same strains continuously in the same child.
Strain identification and drug sensitivity test
The blood culture bottles were placed on the instrument for incubation. The positive samples were transferred to the culture plate and incubated at 37 ℃ for 18–24 h (5% CO2). The colonies were identified using a mass spectrometer. The drug sensitivity test used both the automatic bacterial detection and analysis system and the Kirby–Bauer (KB) method. The results were evaluated according to the latest standards of the Clinical Laboratory Standardization Association. Extended-spectrum β-lactamases (ESBLs) were detected using an automatic bacterial detection and analysis system. The judgment results were derived from its expert system. The quality control strains, Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 27853), Staphylococcus aureus (ATCC 25923 and ATCC 29213), Enterococcus faecalis (ATCC 29212), and Streptococcus pneumoniae (ATCC 49619), were purchased from the Clinical Testing Center of the National Health Commission.
Data analysis
SPSS version 20.0 and WHONET 5.6 (WHO Collaborating Centre for Surveillance of Antimicrobial Resistance, Boston, MA, USA) were used to analyze the data. The data are expressed as means and standard deviations (\(\overline{\mathrm x}\) ± SD). Frequency data are expressed as number of cases (n) and rates (%). The t-test and χ2 test were performed for univariate analysis. Binary logistic regression analysis was used in the multivariable analysis. Statistical significance was set at P < 0.05.
Results
Basic clinical information
Of the included patients (N = 281), 167 were males (59.4%; aged 3.8 ± 4.4 years) and 114 were females (40.6%; aged 3.9 ± 4.1 years). The male-to-female ratio was 1.5:1. The most common diseases were hematological malignancies (n = 74, 26.3%), respiratory system diseases (n = 57, 20.3%), heart diseases (n = 51, 18.2%), and central nervous system diseases (n = 39, 13.9%). Pneumonia and central nervous system infections are the most common hospital-acquired infections, while other infections such as urinary tract infections and phlebitis are less common. Of the included patients, 243 were in the single-infection group and 38 were in the mixed-infection group. Children in both groups had similar ages (3.7 ± 4.2 vs 4.6 ± 5.0, t = 1.095, P = 0.279). There were 259 patients in the survival group and 22 in the death group.
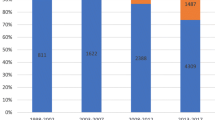
Annual distribution of pathogenic bacteria [n (%)]
As depicted in Fig. 1, Gram-positive bacteria constitute the primary pathogens causing BSI in children admitted to the ICU, consistently exhibiting a higher positive rate compared to Gram-negative bacteria and fungi. Coagulase-negative Staphylococcus (CoNS) is the most common BSI in children (see Supplementary materials Table S1 for details). The positive rates of Gram-positive or Gram-negative pathogens and fungi each year are shown in Fig. 2.
Comparative analysis of clinical characteristics in ICU with BSI [n (%)]
As shown in Table 1, the proportion of male patients aged between 0 and < 3 years with common diseases, such as hematological malignancy, respiratory system disease, heart disease, and central nervous system disease, was higher. Further analysis of mortality risk factors showed a statistically significant difference (P < 0.05) in age, length of hospitalization, primary disease, and pathogen type.
Multivariable analysis of mortality risk factors among children with BSI
To further explore the meaningful indicators of univariate analysis in Table 1, a multi-factor analysis was performed on the mortality risk factors in children with BSI in the ICU (Table 2).
Analysis of resistance of major Gram-positive bacteria to commonly used antibiotics
The resistance rates of Staphylococcus to penicillin and erythromycin were high. None of the detected Staphylococcus strains were resistant to quinuputin/dafopratin, linezolid, vancomycin, teicoplanin, or tigecycline. Table 3 shows the drug resistance analysis of the main Gram-positive bacteria to common antibiotics.
Analysis of resistance of the main Gram-negative bacteria to common antibiotics [n (%)]
Compared with Escherichia coli, Klebsiella pneumoniae showed lower rates of resistance to aztreonam, cefuroxime, cefotaxime, ceftriaxone, ciprofloxacin, levofloxacin, gentamycin, and compound sulfamethoxazole. The resistance rates of Acinetobacter baumannii and Pseudomonas aeruginosa to antibiotics are shown in Table 4.
Detection of MDR bacteria
As shown in Table 5, 27 common MDR bacteria were isolated, including 23 strains of carbapenem-resistant Gram-negative bacteria and four strains of methicillin-resistant Staphylococcus aureus (MRSA). Vancomycin-resistant Enterococcus (VRE) strains were not detected.
Discussion
BSI can manifest as bacteremia or progress to sepsis, a common occurrence among critically ill children in the ICU. The incidence of BSI has increased in recent years. Children might exhibit only transient infection symptoms, while others may develop severe sepsis and shock, often leading to a poor prognosis. Blood culture is currently the gold standard for the diagnosis of BSI, and antibiotics can be selected based on the results of bacterial culture and drug sensitivity in clinical practice (Soedarmono et al. 2022). It has been reported that in-hospital mortality caused by severe BSI is as high as 30–60%, which exceeds the total mortality caused by breast cancer, acquired immunodeficiency syndrome, and prostate cancer (Martínez Pérez-Crespo et al. 2021). Every hour that treatment is delayed increases the mortality of children by 7.6% (Kumar et al. 2006). International guidelines suggest that effective antibiotics should be injected intravenously within 1 h of sepsis diagnosis (Dellinger et al. 2013). Therefore, it is necessary to summarize and analyze the pathogen distribution, related risk factors, and drug sensitivity results of BSI in ICU children to help clinicians select appropriate empirical treatment plans, improve the prognosis of children with sepsis, and reduce BSI mortality.
In our study, 328 pathogenic strains were isolated, including Gram-positive bacteria (68%, 223/328), Gram-negative bacteria (27.7%, 91/328), and fungi (4.3%, 14/328). The main Gram-positive bacteria were CoNS (47.86%, 157/328), Streptococcus pneumoniae (6.10%, 20/328), and Staphylococcus aureus (4.9%, 16/328). The main Gram-negative bacteria were Escherichia coli (5.8%, 19/328), Acinetobacter baumannii (4.9%, 16/328), Klebsiella pneumoniae (4.6%, 15/328), and Pseudomonas aeruginosa (4.3%, 14/328). The fungi type identified was mainly Candida parapsilosis (3.1%, 10/328). As shown in Fig. 1, the positive blood culture rates from 2016 to 2021 are 5.6%, 3%, 3.8%, 3.1%, 4.6%, and 4.3%, respectively. Gram-positive bacteria, represented by CoNS, are the main pathogens that cause BSI in the ICU, and their rate has always been higher than that of Gram-negative bacteria and fungi. A study comparing the pathogens of BSI between children and adults in the ICU found that most adults had Gram-negative bacteria, while the children had CoNS (Zhang et al. 2021). An increasing number of studies have shown that cases of BSI caused by Gram-positive bacteria are on the rise (Santella et al. 2020; Wang et al. 2021; Dambroso-Altafini et al. 2022). In contrast, some studies have shown that Gram-negative bacteria are the main pathogens (Amanati et al. 2021; Zain et al. 2022). Variations in BSI pathogen detection could be attributed to factors, such as timing, geographical location, and study objective. As such, the results solely reflect the situation within the research institution during a specific period. Additionally, similar to previous studies, the most common Gram-negative bacterium causing BSI in this study was Escherichia coli (Zain et al. 2022; Hu et al. 2022). In recent years, the incidence of BSI caused by fungi has increased, with Candida being the most common fungi (Lee et al. 2021). In our study, however, the fungal infection rate in children was lower compared with bacterial infections, and Candida parapsilosis emerged as the main infectious pathogen.
Among the 281 patients with BSI, 243 were infected by one pathogen (86.5%, 243/281) and 38 were infected by mixed pathogens (13.5%, 38/281), which is close to the mixed infection rate reported in the previous studies (6–13%) (Kiani et al. 1979; Rello et al. 1993; Lin et al. 2010). In the mixed-infection group, dual infections with two pathogens constituted 11.03%, triple-pathogen infections accounted for 1.8%, and quadruple-pathogen infections accounted for 0.7%. When comparing the clinical data of children in the single and mixed-infection groups, we found that there was a significant difference in the length of hospitalization between the two groups (P < 0.05). Mixed infections are the most complex and serious type of infection associated with sepsis. The treatment efficacy is suboptimal and the prognosis is often poor (Lin et al. 2010). Early diagnoses, along with timely and effective antibiotic therapy, are keys to improving the prognosis of pediatric patients with sepsis.
To further explore the risk factors for death from BSI, 281 children with BSI in the ICU were divided into a survival group (n = 259) and a death group (n = 22). The length of hospitalization, age, primary diseases, and pathogen type were significantly different between the two groups (P < 0.05). Binary logistic regression analysis revealed that the primary disease, notably hematological malignancies, was an independent risk factor for mortalities (Table 2). Of the 281 children diagnosed with BSI in the ICU, 22 died, resulting in a mortality rate of 7.8%. The mortality rate among the children in the mixed-infection group was 7.89% (3/38), which was slightly higher than that in the single-infection group (7.82% (19/243)). Initially, most mixed infections are considered single-pathogen infections, leading to inadequate empirical drug treatment. This inadequacy exacerbates the patient’s conditions, prolongs hospital stay, and increases mortality rates (Shen et al. 2015). Therefore, it is critical to identify children with a high risk of mixed infections, ascertain the suspected source of infection, and promptly identify the predominant pathogens. This will help understand the antimicrobial resistance patterns within local medical institutions.
In order to remind doctors to understand the current situation of pathogen resistance to certain antibiotics in the local area and to use antibiotics reasonably, we have conducted statistics on the resistance of major pathogens in recent years. The most common Gram-positive bacteria include Staphylococcus epidermidis, Staphylococcus haemolyticus, Streptococcus pneumoniae, Staphylococcus aureus, and Enterococcus faecium. As shown in Table 3, the resistance rates of Staphylococcus to penicillin and erythromycin were high. Vancomycin-resistant strains have also been reported (Fournier et al. 2013). None of the Gram-positive bacteria were resistant to linezolid, vancomycin, teicoplanin, or tigecycline in our study. CoNS belong to the normal flora of the human skin and mucosal tissue. Numerous reports highlight CoNS’s association with infectious diseases, particularly catheter-related BSI (May et al. 2014; Matarrese et al. 2021). Most CoNS strains are methicillin-resistant coagulase-negative Staphylococcus (MRCoNS). In total, 61 strains of methicillin-resistant coagulase-negative Staphylococcus epidermidis (84.7%, 61/72) and 24 strains of methicillin-resistant coagulase-negative Staphylococcus haemolyticus (92.3%, 24/26) were isolated in this study, both of which showed multiple drug resistance. This is consistent with results from a previous study (Peng et al. 2021). Compared with Staphylococcus epidermidis and Staphylococcus haemolyticus, Staphylococcus aureus showed a lower drug resistance rate to some β-lactamides and quinolone antibiotics, and they were 100% sensitive to compound sulfamethoxazole, gentamicin, and tigecycline. Four strains of MRSA and five strains of Staphylococcus aureus with positive D results were detected. MRSA is resistant to all β-lactamide antibiotics. Streptococcus pneumoniae was more than 95% resistant to macrolide antibiotics (erythromycin and clindamycin) and more than 50% resistant to tetracycline, compound sulfamethoxazole, and quinuputin/dafopratin. However, the drug resistance rate to amoxicillin was low (only 10%). Enterococcus faecium was 100% sensitive to quinuputin/dafopratin, linezolid, vancomycin, teicoplanin, and tigecycline, consistent with a recent study (Tian 2022). The resistance rate of Enterococcus faecium to penicillin was as high as 90%, whereas the resistance rate to some quinolone antibiotics was low (30–40%). In our study, the resistance rate of the isolated Gram-positive bacteria to quinolones (ciprofloxacin, levofloxacin, and moxifloxacin) and aminoglycoside antibiotics (gentamicin) was lower than that of other antibiotics, possibly due to the influence of quinolones on bone development, the nephrotoxicity, and ototoxicity of aminoglycoside antibiotics and their reduced use in children.
Of the 22 children who died during the study period, 14 were infected with Gram-negative bacteria (63.6%, 14/22). This highlights the critical need to focus on children infected with Gram-negative bacteria in ICUs. Studies have shown that BSI caused by Gram-negative bacteria is an independent risk factor for high mortality in ICU (Dat et al. 2018). Ninety-one strains of Gram-negative bacteria were isolated, including Escherichia coli, Acinetobacter baumannii, Klebsiella pneumoniae, and Pseudomonas aeruginosa. The drug sensitivity results for the four bacterial strains are presented in Table 4. The composition ratios of the ESBLs-producing strains of Escherichia coli and Klebsiella pneumoniae were 57.9% (11/19) and 53.3% (8/15), respectively. However, the drug resistance rate of Klebsiella pneumoniae to imipenem was higher than that of Escherichia coli (20% vs.5.6%). Studies have shown that the incidence of BSI caused by carbapenem-resistant Klebsiella pneumoniae (CRKP) is increasing (Stein et al. 2019; Guo et al. 2023). According to data from the National Drug Resistance Monitoring Network (http://www.carss.cn/), the isolation rate of CRKP among children in China increased from 3.0 to 20.9% from 2005 to 2017, which was significantly higher than that among adults (Wang et al. 2020). Compared with Escherichia coli, Klebsiella pneumoniae has a lower rate of resistance to aztreonam, cefuroxime, cefotaxime, ceftriaxone, ciprofloxacin, levofloxacin, gentamycin, and cotrimoxazole. The resistance rate of Acinetobacter baumannii to various antibiotics was higher than 75%, whereas the resistance rates to levofloxacin and minocycline were lower at 43.8% and 37.5%, respectively. These results highlight the concerning drug resistance of Acinetobacter baumannii, revealing a limited array of effective drug options. A combination of tegacycline-based drugs for the treatment of severe infections caused by Acinetobacter baumannii is the more commonly used regimen. Recently, our research group conducted a study on the carbapenem resistance and virulence of Acinetobacter baumannii and analyzed the cause of its multiple drug resistance (Zhu et al. 2022). Pseudomonas aeruginosa is sensitive to commonly used clinical anti-pseudomonas drugs, with a resistance rate of 21.4% to aztreonam, 21.4% to cefoperazone/sulbactam, 7.1% to piperacillin/tazobactam, 7.1% to ceftazidime, and 21.4% to cefepime. Pseudomonas aeruginosa was 100% resistant to quinolones (ciprofloxacin and levofloxacin) and aminoglycosides (gentamicin, tobramycin, and amikacin).
Among the 328 pathogenic bacteria identified in this study, 27 common MDR bacteria were isolated, including 13 strains of carbapenem-resistant Acinetobacter baumannii (CRAB), six strains of carbapenem-resistant Pseudomonas aeruginosa (CRPA), three strains of carbapenem-resistant Klebsiella pneumoniae (CRKP), one strain of carbapenem-resistant Escherichia coli (CREO), and four strains of MRSA. VRE strains were not detected. More MDR bacteria strains were detected in 2021. In this study, the proportion of CRAB is high (48.2%; 13/27), which is consistent with another study (Bedenić et al. 2023). The proportion of patients with CRPA was the second highest, accounting for 22.2% (6/27). Among the 22 children who died, four were infected with CRAB and two with CRPA. Carbapenem-resistant Enterobacter (CRE) can be found in the urine, respiratory tract, feces, blood, and other samples (Kotb et al. 2020; Sexton et al. 2022; Xiong et al. 2023). Four CRE strains were identified in this study. These results underscore the need to focus on BSI caused by CR bacteria. Studies have demonstrated that the main cause of the resistance of pathogenic bacteria to carbapenem antibiotics in children is the production of metalloenzymes (class B) (Buys et al. 2016). Currently, the most effective antibiotic combination against CRE is polymyxin coupled with tigecycline (Vanegas et al. 2016). However, tigecycline is rarely used in children because it causes tooth staining. Polymyxins alone are a relatively safe treatment for children. Further analysis of the clinical treatment effects, such as the length of hospitalization and whether there is improvement in these MDR-infected patients, is necessary. Among the 22 children who died, 17 had hematological malignancies (> 50% of the proportion). Therefore, it is essential to focus on BSI in children with hematological malignancies.
In summary, the pathogens causing BSI in children admitted to the ICU in the past 6 years are mainly Gram-positive bacteria such as CoNS. Escherichia coli is the most common Gram-negative bacterium. As such, continuous monitoring of blood cultures in critically ill children with BSI is necessary. This entails focusing on detecting MDR bacteria, improving antibiotic application and management, and enhancing hospital infection prevention and control measures. This study has some limitations. Due to the small variation in the number of pathogens detected each year, an analysis of the annual changes in antimicrobial resistance could not be performed. We plan to expand the study period in the next study to more accurately analyze the changes in drug resistance rate.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- KB :
-
Kirby–Baue
- ICU :
-
Intensive care unit
- MDR :
-
Multidrug resistant
- BSI :
-
Bloodstream infection
- VRE :
-
Vancomycin-resistant Enterococcus
- CoNS :
-
Coagulase-negative staphylococci
- ESBLs :
-
Extended-spectrum β-lactamases
- CRE :
-
Carbapenem-resistant enterobacter
- CREO :
-
Carbapenem-resistant Escherichia coli
- CRKP :
-
Carbapenem-resistant Klebsiella pneumoniae
- CRAB :
-
Carbapenem-resistant Acinetobacter baumannii
- CRPA :
-
Carbapenem-resistant Pseudomonas aeruginosa
- MRSA :
-
Methicillin-resistant Staphylococcus aureus
- MRCoNS :
-
Methicillin-resistant coagulase-negative staphylococcus
References
Amanati A, Sajedianfard S, Khajeh S, Ghasempour S, Mehrangiz S, Nematolahi S, Shahhosein Z (2021) Bloodstream infections in adult patients with malignancy, epidemiology, microbiology, and risk factors associated with mortality and multi-drug resistance. BMC Infect Dis 21(1):636. https://doi.org/10.1186/s12879-021-06243-z
Bammigatti C, Doradla S, Belgode HN, Kumar H, Swaminathan RP (2017) Healthcare associated infections in a resource limited setting. J Clin Diagn Res: JCDR 11(1):OC01–OC04. https://doi.org/10.7860/JCDR/2017/23076.9150
Bassetti M, De Waele JJ, Eggimann P, Garnacho-Montero J, Kahlmeter G, Menichetti F, Nicolau DP, Paiva JA, Tumbarello M, Welte T, Wilcox M, Zahar JR, Poulakou G (2015) Preventive and therapeutic strategies in critically ill patients with highly resistant bacteria. Intensive Care Med 41(5):776–795. https://doi.org/10.1007/s00134-015-3719-z
Bedenić B, Bratić V, Mihaljević S, Lukić A, Vidović K, Reiner K, Schöenthaler S, Barišić I, Zarfel G, Grisold A (2023) Multidrug-resistant bacteria in a COVID-19 hospital in Zagreb. Pathogens (basel, Switzerland) 12(1):117. https://doi.org/10.3390/pathogens12010117
Buys H, Muloiwa R, Bamford C, Eley B (2016) Klebsiella pneumoniae bloodstream infections at a South African children’s hospital 2006–2011, a cross-sectional study. BMC Infect Dis 16(1):570. https://doi.org/10.1186/s12879-016-1919-y
Centers for disease control and prevention/national healthcare safety network. CDC/NHSN Surveillance Definitions for Specific Types of Infections. January 2017
Dambroso-Altafini D, Menegucci TC, Costa BB, Moreira RRB, Nishiyama SAB, Mazucheli J, Tognim MCB (2022) Routine laboratory biomarkers used to predict Gram-positive or Gram-negative bacteria involved in bloodstream infections. Sci Rep 12(1):15466. https://doi.org/10.1038/s41598-022-19643-1
Dat VQ, Long NT, Hieu VN, Phuc NDH, Kinh NV, Trung NV, van Doorn HR, Bonell A, Nadjm B (2018) Clinical characteristics, organ failure, inflammatory markers and prediction of mortality in patients with community acquired bloodstream infection. BMC Infect Dis 18(1):535. https://doi.org/10.1186/s12879-018-3448-3
Dellinger P, Levy MM, Rhodes A et al (2013) Action to save patients with sepsis: International Guidelines for the Treatment of Severe Sepsis and Septic Shock 2012[S]. Crit Care Med 41(2):580–586
Fournier PE, Drancourt M, Colson P, Rolain JM, La Scola B, Raoult D (2013) Modern clinical microbiology: new challenges and solutions. Nat Rev Microbiol 11(8):574–585. https://doi.org/10.1038/nrmicro3068
Gouel-Cheron A, Swihart BJ, Warner S, Mathew L, Strich JR, Mancera A, Follmann D, Kadri SS (2022) Epidemiology of ICU-onset bloodstream infection: prevalence, pathogens, and risk factors among 150,948 ICU patients at 85 U.S. hospitals. Crit Care Med 50(12):1725–1736. https://doi.org/10.1097/CCM.0000000000005662
Guo, Y., Liu, F., Zhang, Y., Wang, X., Gao, W., Xu, B., Li, Y., & Song, N. (2023). Virulence, antimicrobial resistance, and molecular characteristics of carbapenem-resistant Klebsiella pneumoniae in a hospital in Shijiazhuang City from China. Int Microbiol. https://doi.org/10.1007/s10123-023-00357-x. Advance online publication. https://doi.org/10.1007/s10123-023-00357-x
Hu F, Yuan L, Yang Y, Xu Y, Huang Y, Hu Y, Ai X, Zhuo C, Su D, Shan B, Du Y, Yu Y, Lin J, Sun Z, Chen Z, Xu Y, Zhang X, Wang C, He L, Ni Y, … Zhang Y. (2022). A multicenter investigation of 2,773 cases of bloodstream infections based on China antimicrobial surveillance network (CHINET). Front Cell Infect Microbiol 12:1075185. https://doi.org/10.3389/fcimb.2022.1075185
Kiani D, Quinn EL, Burch KH, Madhavan T, Saravolatz LD, Neblett TR (1979) The increasing importance of polymicrobial bacteremia. JAMA 242(10):1044–1047
Kotb S, Lyman M, Ismail G, Abd El Fattah M, Girgis SA, Etman A, Hafez S, El-Kholy J, Zaki MES, Rashed HG, Khalil GM, Sayyouh O, Talaat M (2020) Epidemiology of carbapenem-resistant Enterobacteriaceae in Egyptian intensive care units using National Healthcare-associated Infections Surveillance Data, 2011–2017. Antimicrob Resist Infect Control 9(1):2. https://doi.org/10.1186/s13756-019-0639-7
Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, Suppes R, Feinstein D, Zanotti S, Taiberg L, Gurka D, Kumar A, Cheang M (2006) Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 34(6):1589–1596. https://doi.org/10.1097/01.CCM.0000217961.75225.E9
Lee Y, Puumala E, Robbins N, Cowen LE (2021) Antifungal drug resistance: molecular mechanisms in Candida albicans and beyond. Chem Rev 121(6):3390–3411. https://doi.org/10.1021/acs.chemrev.0c00199
Lin JN, Lai CH, Chen YH, Chang LL, Lu PL, Tsai SS, Lin HL, Lin HH (2010) Characteristics and outcomes of polymicrobial bloodstream infections in the emergency department: a matched case-control study. Acad Emerg Med off J Soc Acad Emerg Med 17(10):1072–1079. https://doi.org/10.1111/j.1553-2712.2010.00871.x\
Markwart R, Saito H, Harder T, Tomczyk S, Cassini A, Fleischmann-Struzek C, Reichert F, Eckmanns T, Allegranzi B (2020) Epidemiology and burden of sepsis acquired in hospitals and intensive care units: a systematic review and meta-analysis. Intensive Care Med 46(8):1536–1551. https://doi.org/10.1007/s00134-020-06106-2
Marsillio LE, Ginsburg SL, Rosenbaum CH, Coffin SE, Naim MY, Priestley MA, Srinivasan V (2015) Hyperglycemia at the time of acquiring central catheter-associated bloodstream infections is associated with mortality in critically Ill children. Pediatr Crit Care Med 16(7):621–628. https://doi.org/10.1097/PCC.0000000000000445
Martínez Pérez-Crespo PM, López-Cortés LE, Retamar-Gentil P, García JFL, Vinuesa García D, León E, Calvo JMS, Galán-Sánchez F, Natera Kindelan C, Del Arco Jiménez A, Sánchez-Porto A, Herrero Rodríguez C, Becerril Carral B, Molina IMR, Iglesias JMR, Pérez Camacho I, Guzman García M, López-Hernández I, Rodríguez-Baño J, PROBAC REIPI/GEIH-SEIMC/SAEI Group (2021) Epidemiologic changes in bloodstream infections in Andalucía (Spain) during the last decade. Clin Microbiol Infect 27(2):283.e9-283.e16. https://doi.org/10.1016/j.cmi.2020.05.015
Matarrese AN, Ivulich DI, Cesar G, Alaniz F, Ruiz JJ, Osatnik J (2021) Análisis epidemiológico de bacteriemias asociadas a catéter en una terapia intensiva médico-quirúrgica [Epidemiological analysis of catheter-related bloodstream infections in medical-surgical intensive care units]. Medicina 81(2):159–165
May L, Klein EY, Rothman RE, Laxminarayan R (2014) Trends in antibiotic resistance in coagulase-negative staphylococci in the United States, 1999 to 2012. Antimicrob Agents Chemother 58(3):1404–1409. https://doi.org/10.1128/AAC.01908-13
Ministry of health of the People’s Republic of China (2003) Diagnostic criteria for nosocomial infection (Trial)[M]. Modern Pract Med 15(7):460–464
Peng ZL, Jiang Y, Jia LJ et al (2021) Analysis of drug resistance and prognosis of bloodstream infection pathogens. Clin Meta Anal 36(8):724–729
Rello J, Quintana E, Mirelis B, Gurguí M, Net A, Prats G (1993) Polymicrobial bacteremia in critically ill patients. Intensive Care Med 19(1):22–25. https://doi.org/10.1007/BF01709273
Santella B, Folliero V, Pirofalo GM, Serretiello E, Zannella C, Moccia G, Santoro E, Sanna G, Motta O, De Caro F, Pagliano P, Capunzo M, Galdiero M, Boccia G, Franci G (2020) Sepsis-A retrospective cohort study of bloodstream infections. Antibiotics (basel, Switzerland) 9(12):851. https://doi.org/10.3390/antibiotics9120851
Schwab F, Geffers C, Behnke M, Gastmeier P (2018) ICU mortality following ICU-acquired primary bloodstream infections according to the type of pathogen: a prospective cohort study in 937 Germany ICUs (2006–2015). PLoS ONE 13(3):e0194210. https://doi.org/10.1371/journal.pone.0194210
Sexton ME, Bower C, Jacob JT (2022) Risk factors for isolation of carbapenem-resistant Enterobacterales from normally sterile sites and urine. Am J Infect Control 50(8):929–933. https://doi.org/10.1016/j.ajic.2021.12.007
Shen FC, Xie D, Han QP, Zeng HK, Deng XY (2015) Pathogen characteristics of bloodstream infection in ICU and risk factors analysis of mixed blood flow infection. China Crit Care Med 27(9):718–723. https://doi.org/10.3760/cma.j.issn.2095-4352.2015.09.004
Shime N, Kawasaki T, Saito O, Akamine Y, Toda Y, Takeuchi M, Sugimura H, Sakurai Y, Iijima M, Ueta I, Shimizu N, Nakagawa S (2012) Incidence and risk factors for mortality in paediatric severe sepsis: results from the national paediatric intensive care registry in Japan. Intensive Care Med 38(7):1191–1197. https://doi.org/10.1007/s00134-012-2550-z
Soedarmono P, Diana A, Tauran P, Lokida D, Aman AT, Alisjahbana B, Arlinda D, Tjitra E, Kosasih H, Merati KTP, Arif M, Gasem MH, Susanto NH, Lukman N, Sugiyono RI, Hadi U, Lisdawati V, Tchos KGF, Neal A, Karyana M (2022) The characteristics of bacteremia among patients with acute febrile illness requiring hospitalization in Indonesia. PLoS ONE 17(9):e0273414. https://doi.org/10.1371/journal.pone.0273414
Stein C, Vincze S, Kipp F, Makarewicz O, Al Dahouk S, Pletz MW (2019) Carbapenem-resistant Klebsiella pneumoniae with low chlorhexidine susceptibility. Lancet Infect Dis 19(9):932–933. https://doi.org/10.1016/S1473-3099(19)30427-X
Tian Q (2022) Analysis of pathogenic bacteria distribution and drug resistance in patients with bloodstream infection in ICU. Explor Rational Drug Use China 19(2):29–35
Timsit JF, Ruppé E, Barbier F, Tabah A, Bassetti M (2020) Bloodstream infections in critically ill patients: an expert statement. Intensive Care Med 46(2):266–284. https://doi.org/10.1007/s00134-020-05950-6
Tran K, Bell C, Stall N, Tomlinson G, McGeer A, Morris A, Gardam M, Abrams HB (2017) The effect of hospital isolation precautions on patient outcomes and cost of care: a multi-site, retrospective, propensity score-matched cohort study. J Gen Intern Med 32(3):262–268. https://doi.org/10.1007/s11606-016-3862-4
Vanegas JM, Parra OL, Jiménez JN (2016) Molecular epidemiology of carbapenem resistant gram-negative bacilli from infected pediatric population in tertiary-care hospitals in Medellín, Colombia: an increasing problem. BMC Infect Dis 16(1):463. https://doi.org/10.1186/s12879-016-1805-7
Wang B, Pan F, Wang C, Zhao W, Sun Y, Zhang T, Shi Y, Zhang H (2020) Molecular epidemiology of Carbapenem-resistant Klebsiella pneumoniae in a paediatric hospital in China. Int J Infect Dis 93:311–319. https://doi.org/10.1016/j.ijid.2020.02.009
Wang C, Hao W, Yu R, Wang X, Zhang J, Wang B (2021) Analysis of pathogen distribution and its antimicrobial resistance in bloodstream infections in hospitalized children in East China, 2015–2018. J Trop Pediatr 67(1):fmaa077. https://doi.org/10.1093/tropej/fmaa077
Xie J, Li S, Xue M, Yang C, Huang Y, Chihade DB, Liu L, Yang Y, Qiu H (2020) Early- and late-onset bloodstream infections in the intensive care unit: a retrospective 5-year study of patients at a university hospital in China. J Infect Dis 221(Suppl 2):S184–S192. https://doi.org/10.1093/infdis/jiz606
Xiong Z, Zhang C, Sarbandi K, Liang Z, Mai J, Liang B, Cai H, Chen X, Gao F, Lan F, Liu X, Liu S, Zhou Z (2023) Clinical and molecular epidemiology of carbapenem-resistant Enterobacteriaceae in pediatric inpatients in South China. Microbiol Spectrum e0283923. Advance online publication. https://doi.org/10.1128/spectrum.02839-23
Yan G, Liu J, Chen W, Chen Y, Cheng Y, Tao J, Cai X, Zhou Y, Wang Y, Wang M, Lu G (2021) Metagenomic next-generation sequencing of bloodstream microbial cell-free nucleic acid in children with suspected sepsis in pediatric intensive care unit. Front Cell Infect Microbiol 11:665226. https://doi.org/10.3389/fcimb.2021.665226
Zain OM, Elsayed MY, Abdelkhalig SM, Abdelaziz M, Ibrahim SY, Bashir T, Hamadalnil Y (2022) Bloodstream infection in cancer patients; susceptibility profiles of the isolated pathogens, at Khartoum Oncology Hospital. Sudan African Health Sci 22(4):70–76. https://doi.org/10.4314/ahs.v22i4.10
Zhang Y, Zhou J, Cao T, Zheqian Li (2021) Comparison of pathogenic bacteria distribution, drug resistance and clinical characteristics of bloodstream infection between children and adults admitted to intensive care unit. Shandong Med 61(12):4–10. https://doi.org/10.3969/j.issn.1002-266X.2021.12.005
Zhang Y, Cao B, Cao W, Miao H, Wu L (2022) Clinical characteristics and death risk factors of severe sepsis in children. Comput Math Methods Med 2022:4200605. https://doi.org/10.1155/2022/4200605
Zhu S, Kang Y, Wang W, Cai L, Sun X, Zong Z (2019) The clinical impacts and risk factors for non-central line-associated bloodstream infection in 5046 intensive care unit patients: an observational study based on electronic medical records. Crit Care (london, England) 23(1):52. https://doi.org/10.1186/s13054-019-2353-5
Zhu Y, Zhang X, Wang Y, Tao Y, Shao X, Li Y, Li W (2022) Insight into Carbapenem resistance and virulence of Acinetobacter baumannii from a children’s medical centre in eastern China. Ann Clin Microbiol Antimicrob 21(1):47. https://doi.org/10.1186/s12941-022-00536-0
Acknowledgements
We thank the staff from the Department of Clinical Laboratory, Children’s Hospital of Soochow University, who took part in the study.
Funding
This study was supported by grants from the Special Foundation for National Science and Technology Basic Research Program of China (2019FY101200), the High-level Innovative and Entrepreneurial Talents Introduction Program of Jiangsu Province (2020–30191), the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (20KJB310012), the Medical Research Project of Jiangsu Commission of Health (M2020027), the “National Tutorial System” Project for Suzhou Young Health Talents (Qngg2022011), the Science and Technology Program of Suzhou (SYS2020163, SYSD2019120, SLC201904), and the Youth Science and Program of Suzhou (KJXW2023025).
Author information
Authors and Affiliations
Contributions
HJ S and L D conceived the study and designed the experiments. Y L and XJ S provide financial support. HJ S, X Z, and YY G collected and analyzed the data. YZ W and L D interpreted the results. HJ S and X Z drafted the manuscript, and all authors critically revised the manuscript for intellectual content and read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
It was reviewed and approved by the Medical Ethics Committee of the Children's Hospital of Soochow University (Ethics batch number: 2021CS158). Informed consent was obtained from all subjects and/or their legal guardian(s). All methods were performed in accordance with the relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Disclaimer
The views, opinions, assumptions, or any other information set out in this article are solely those of the authors and should not be attributed to the funders or any other person connected with the funders.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary File 1
(DOCX 25.1 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shao, H., Zhang, X., Li, Y. et al. Epidemiology and drug resistance analysis of bloodstream infections in an intensive care unit from a children’s medical center in Eastern China for six consecutive years. Int Microbiol (2024). https://doi.org/10.1007/s10123-024-00481-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10123-024-00481-2