Abstract
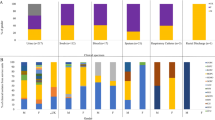
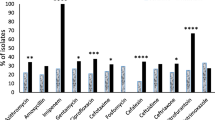
Morganella morganii is a bacterium belonging to the normal intestinal microbiota and the environment; however, in immunocompromised individuals, this bacterium can become an opportunistic pathogen, causing a series of diseases, both in hospitals and in the community, being urinary tract infections more prevalent. Therefore, the objective of this study was to evaluate the prevalence, virulence profile, and resistance to antimicrobials and the clonal relationship of isolates of urinary tract infections (UTI) caused by M. morganii, both in the hospital environment and in the community of the municipality of Londrina-PR, in southern Brazil, in order to better understand the mechanisms for the establishment of the disease caused by this bacterium. Our study showed that M. morganii presents a variety of virulence factors in the studied isolates. Hospital strains showed a higher prevalence for the virulence genes zapA, iutA, and fimH, while community strains showed a higher prevalence for the ireA and iutA genes. Hospital isolates showed greater resistance compared to community isolates, as well as a higher prevalence of multidrug-resistant (MDR) and extended-spectrum beta lactamase (ESBL)-producing isolates. Several M. morganii isolates from both sources showed high genetic similarity. The most prevalent plasmid incompatibility groups detected were FIB and I1, regardless of the isolation source. Thus, M. morganii isolates can accumulate virulence factors and antimicrobial resistance, making them a neglected opportunistic pathogen.




Similar content being viewed by others
References
Adeolu M, Alnajar S, Naushad S, S Gupta R (2016) Genome-based phylogeny and taxonomy of the “Enterobacteriales”: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int J Syst Evol Microbiol 66:5575–5599. https://doi.org/10.1099/ijsem.0.001485
Andersson DI, Balaban NQ, Baquero F, Courvalin P, Glaser P, Gophna U, Kishony R, Molin S, Tønjum T (2020) Antibiotic resistance: turning evolutionary principles into clinical reality. FEMS Microbiol Rev 44:171–18810 1093/femsre/fuaa001
Arlet AP (1991) Construction by polymerase chain reaction and use of intragenic DNA probes for three main types of transferable β-lactamase (TEM, SHV, CARB). FEMS Microbiol Lett. https://doi.org/10.1111/j.1574-6968.1991.tb04833.x
Arredondo-García JL, Figueroa-Damián R, Rosas A, Jáuregui A, Corral M, Costa A, Cardeñosa-Guerra O (2004) Comparison of short-term treatment regimen of ciprofloxacin versus long-term treatment regimens of trimethoprim/sulfamethoxazole or norfloxacin for uncomplicated lower urinary tract infections: a randomized, multicentre, open-label, prospective study. J Antimicrob Chemother. https://doi.org/10.1093/jac/dkh414
Bandy A (2020) Ringing bells: Morganella morganii fights for recognition. Public Health 182:45–50. https://doi.org/10.1016/j.puhe.2020.01.016
Bilge SS, Clausen CR, Lau W, Moseley SL (1989) Molecular characterization of a fimbrial adhesin, F1845, mediating diffuse adherence of diarrhea-associated Escherichia coli to HEp-2 cells. J Bacteriol. https://doi.org/10.1128/jb.171.8.4281-4289.1989
Caneiras C, Lito L, Melo-Cristino J, Duarte A (2019) Community-and hospital-acquired Klebsiella pneumoniae urinary tract infections in Portugal: virulence and antibiotic resistance. Microorg. https://doi.org/10.3390/microorganisms7050138
Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ (2005) Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. https://doi.org/10.1016/j.mimet.2005.03.018
Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P (2007) Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J Antimicrob Chemother. https://doi.org/10.1093/jac/dkm204
Chen X, Zhang W, Pan W, Yin J, Pan Z, Gao S, Jiao X (2012a) Prevalence of qnr, aac (6′)-Ib-cr, qepA, and oqxAB in Escherichia coli isolates from humans, animals, and the environment. Antimicrob Agents Chemother. https://doi.org/10.1128/AAC.06191-11
Chen YP, Lei CW, Kong LH, Zeng JX, Zhang XZ, Liu BH, Wang HN (2018) Tn 6450, a novel multidrug resistance transposon characterized in a Proteus mirabilis isolate from chicken in China. Antimicrob Agents Chemother. https://doi.org/10.1128/AAC.02192-17
Chen Y-T, Peng H-L, Shia W-C, Hsu F-R, Ken C-F, Tsao Y-M, Chen C-H, Liu C-E, Hsieh M-F, Chen H-C, Tang C-Y, Ku T-H (2012b) Whole-genome sequencing and identification of Morganella morganii KT pathogenicity-related genes. BMC Genomics 13(Suppl 7):S4. https://doi.org/10.1186/1471-2164-13-S7-S4
Clinical and Laboratory Standards Instuitte (CLSI) (2021) Performance standards for antimicrobial susceptibility testing, 28th edn. CLSI supplement M100. Wayne, p 32
Cravioto A, Gross RJ, Scotland SM, Rowe B (1979) An adhesive factor found in strains of Escherichia coli belonging to the traditional infantile enteropathogenic serotypes. Current Microbiol. https://doi.org/10.1007/BF02602439
Cyoia PS, Koga VL, Nishio EK, Houle S, Dozois CM, De Brito KCT, Kobayashi RKT (2019) Distribution of ExPEC virulence factors, blaCTX-M, fosA3, and mcr-1 in Escherichia coli isolated from commercialized chicken carcasses. Front Microbiol. https://doi.org/10.3389/fmicb.2018.03254
Davies D (2003) Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. https://doi.org/10.1038/nrd1008
De La Cadena E, Mojica MF, Castillo N, Correa A, Appel TM, García-Betancur JC, Villegas MV (2020) Genomic analysis of CTX-M-group-1-producing extraintestinal pathogenic E. coli (ExPEc) from patients with urinary tract infections (UTI) from Colombia. Antibiot. https://doi.org/10.3390/antibiotics9120899
de Oliveira WD, Barboza MGL, Faustino G, Inagaki WTY, Sanches MS, Kobayashi RKT, Rocha SPD (2021) Virulence, resistance and clonality of Proteus mirabilis isolated from patients with community-acquired urinary tract infection (CA-UTI) in Brazil. Microb Pathog. https://doi.org/10.1016/j.micpath.2020.104642
Di Venanzio G, Stepanenko TM, García VE (2014) Serratia marcescens ShlA pore-forming toxin is responsible for early induction of autophagy in host cells and is transcriptionally regulated by RcsB. Infect Immun. https://doi.org/10.1128/IAI.01682-14
Eberspacher B, Hugo F, Pohl M, Bhakdi S (1990) Functional similarity between the haemolysins of Escherichia coli and Morganella morganii. J Med Microbiol. https://doi.org/10.1099/00222615-33-3-165
Falagas ME, Kavvadia PK, Mantadakis E, Kofteridis DP, Bliziotis IA, Saloustros E, Maraki S, Samonis G (2006) Morganella morganii infections in a general tertiary hospital. Infection 34:315–321. https://doi.org/10.1007/s15010-006-6682-3
Ferjani S, Saidani M, Amine FS, Boutiba-Bem IB (2015) Prevalence and characterization of plasmid-mediated quinolone resistance genes in extended-spectrum β-lactamase-producing Enterobacteriaceae in a Tunisian hospital. Microb Drug Resist. https://doi.org/10.1089/mdr.2014.0053
Gellatly SL, Hancock RE (2013) Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis. https://doi.org/10.1111/2049-632X.12033
Ghavidel M, Gholamhosseini-Moghadam T, Nourian K, Ghazvini K (2020) Virulence factors analysis and antibiotic resistance of uropathogenic Escherichia coli isolated from patients in northeast of Iran. Iran J Microb 12(3):223
Ghosh S, LaPara TM (2007) The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria. ISME J 1:191–203. https://doi.org/10.1038/ismej.2007.31
González GM, Treviño-Rangel RDJ, Campos CL, Villanueva-Lozano H, Bonifaz A, Franco-Cendejas R, Andrade A (2020) Surveillance of antimicrobial resistance in Serratia marcescens in Mexico. New Microbiol 43(1):34–37
Haghighatpanah M, Mojtahedi A (2019) Characterization of antibiotic resistance and virulence factors of Escherichia coli strains isolated from Iranian inpatients with urinary tract infections. Infect Drug Resist. https://doi.org/10.2147/IDR.S219696
Heras J, Domínguez C, Mata E, Pascual V, Lozano C, Torres C, Zarazaga M (2015) GelJ–a tool for analyzing DNA fingerprint gel images. BMC Bioinform. https://doi.org/10.1186/s12859-015-0703-0
Ikeda M, Kobayashi T, Fujimoto F, Okada Y, Higurashi Y, Tatsuno K, Moriya K (2021) The prevalence of the iutA and ibeA genes in Escherichia coli isolates from severe and non-severe patients with bacteremic acute biliary tract infection is significantly different. Gut Pathogens. https://doi.org/10.1186/s13099-021-00429-1
Irrgang A, Hammerl JA, Falgenhauer L, Guiral E, Schmoger S, Imirzalioglu C, Käsbohrer A (2018) Diversity of CTX-M-1-producing E. coli from German food samples and genetic diversity of the blaCTX-M-1 region on IncI1 ST3 plasmids. Vet Microbiol. https://doi.org/10.1016/j.vetmic.2018.06.003
Kader AA, Kumar A (2005) Prevalence and antimicrobial susceptibility of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in a general hospital. Annals Saudi Med. https://doi.org/10.5144/0256-4947.2005.239
Konowalchuk J, Speirs JI, Stavric S (1977) Vero response to a cytotoxin of Escherichia coli. Infect Immun. https://doi.org/10.1128/iai.18.3.775-779.1977
Koronakis V, Cross M, Senior B, Koronakis E, Hughes C (1987) The secreted hemolysins of Proteus mirabilis, Proteus vulgaris, and Morganella morganii are genetically related to each other and to the alpha-hemolysin of Escherichia coli. J Bacteriol. https://doi.org/10.1128/jb.169.4.1509-1515.1987
Kwiecinska-Piróg J, Bogiel T, Skowron K, Wieckowska E, Gospodarek E (2014) Proteus mirabilis biofilm-qualitative and quantitative colorimetric methods-based evaluation. Braz J Microbiol. https://doi.org/10.1590/S1517-83822014000400037
Levine M, Nalin D, Hornick R, Bergquist E, Waterman D, Young C, Rowe B (1978) Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet. https://doi.org/10.1016/S0140-6736(78)90299-4
Leylabadlo HE, Kafil HS, Yousefi M, Aghazadeh M, Asgharzadeh M (2016) Persistent infection with metallo-beta-lactamase and extended spectrum β-lactamase producer Morganella morganii in a patient with urinary tract infection after kidney transplantation. J Nat Sci Biol Med 7:179–181. https://doi.org/10.4103/0976-9668.184707
Li G, Niu X, Yuan S, Liang L, Liu Y, Hu L, Liu J, Cheng Z (2018) Emergence of Morganella morganii subsp. morganii in dairy calves, China. Emerg Microbes Infect 7:172. https://doi.org/10.1038/s41426-018-0173-3
Li Q, Sherwood JS, Logue CM (2007) Characterization of antimicrobial resistant Escherichia coli isolated from processed bison carcasses. J Appl Microbiol. https://doi.org/10.1111/j.1365-2672.2007.03470.x
Liu H, Zhu J, Hu Q, Rao X (2016) Morganella morganii, a non-negligent opportunistic pathogen. Int J Infect Dis 50:10–17. https://doi.org/10.1016/j.ijid.2016.07.006
Livani F, Kabir S (2019) Gram-negative folliculitis caused by Morganella morganii. JAAD Case Rep 5:558–559. https://doi.org/10.1016/j.jdcr.2019.04.021
Manyahi J, Moyo SJ, Tellevik MG, Ndugulile F, Urassa W, Blomberg B, Langeland N (2017) Detection of CTX-M-15 beta-lactamases in Enterobacteriaceae causing hospital-and community-acquired urinary tract infections as early as 2004, in Dar es Salaam, Tanzania. BMC Infect Dis. https://doi.org/10.1186/s12879-017-2395-8
Mazzariol A, Kocsis B, Koncan R, Kocsis E, Lanzafame P, Cornaglia G (2012) Description and plasmid characterization of qnrD determinants in Proteus mirabilis and Morganella morganii. Clin Microbiol Infect. https://doi.org/10.1128/AAC.02192-17
Menard S (2002) Applied logistic regression analysis, 2nd edn. SAGE Publications, Inc, United States of America. https://doi.org/10.4135/9781412983433
Michelim L, Muller G, Zacaria J, Delamare APL, Costa SOPD, Echeverrigaray S (2008) Comparison of PCR-based molecular markers for the characterization of Proteus mirabilis clinical isolates. Braz J Infect Dis. https://doi.org/10.1590/s1413-86702008000500014
Minnullina L, Pudova D, Shagimardanova E, Shigapova L, Sharipova M, Mardanova A (2019) Comparative genome analysis of uropathogenic Morganella morganii strains. Front Cell Infect Microbiol 9:167. https://doi.org/10.3389/fcimb.2019.00167
Morgan H d R (1906) Report XCV. Upon the bacteriology of the summer diarrhoea of infants. Br Med J 1:908–912. https://doi.org/10.1136/bmj.1.2364.908
Mostafavi S, Rostami S, Rezaee Nejad Y, Ataei B, Mobasherizadeh S, Cheraghi A, Shokri D (2021) Antimicrobial resistance in hospitalized patients with community acquired urinary tract infection in Isfahan, Iran. Arch Iran Med. https://doi.org/10.34172/aim.2021.29
Murakami A, Nakamura Y, Torikai K, Tanaka T, Koshiba T, Koshimizu K, Ohigashi H (2000) Inhibitory effect of citrus nobiletin on phorbol ester-induced skin inflammation, oxidative stress, and tumor promotion in mice. Cancer Res 60(18):5059–5066
Naquin E, Soorya H, Oubre C, Rogers S, Boopathy R (2021) Effect of sulfonamide class of antibiotics on a bacterial consortium isolated from an anaerobic digester of a sewage treatment plant. Env Qual Manag. https://doi.org/10.1002/tqem.21709
Nataro JP, Yikang D, Cookson S, Cravioto A, Savarino SJ, Guers LD, Tacket CO (1995) Heterogeneity of enteroaggregative Escherichia coli virulence demonstrated. J Infect Dis. https://doi.org/10.1093/infdis/171.2.465
Pathirana HNKS, De Silva BCJ, Wimalasena SHMP, Hossain S, Heo GJ (2018) Comparison of virulence genes in Proteus species isolated from human and pet turtle. Iran J Vet Res 19(1):48
Patil AB, Nadagir SD, Lakshminarayana S, Syeda FM (2012) Morganella morganii, subspecies morganii, biogroup A: an unusual causative pathogen of brain abscess. J Neurosci Rural Pract 3:370–372. https://doi.org/10.4103/0976-3147.102631
Reuland EA, Al Naiemi N, Kaiser AM, Heck M, Kluytmans JAJW, Savelkoul PHM, Vandenbroucke-Grauls CMJE (2016) Prevalence and risk factors for carriage of ESBL-producing Enterobacteriaceae in Amsterdam. J Antimicrob Chemother. https://doi.org/10.1093/jac/dkv441
Robinson AE, Heffernan JR, Henderson JP (2018) The iron hand of uropathogenic Escherichia coli: the role of transition metal control in virulence. Fut microbiol. https://doi.org/10.2217/fmb-2017-0295
Rocha SP, Pelayo JS, Elias WP (2007) Fimbriae of uropathogenic Proteus mirabilis. FEMS Immunol Med Microbiol. https://doi.org/10.1111/j.1574-695X.2007.00284.x
Russo TA, Carlino UB, Johnson JR (2001) Identification of a new iron-regulated virulence gene, ireA, in an extraintestinal pathogenic isolate of Escherichia coli. Infect Immun. https://doi.org/10.1128/IAI.69.10.6209-6216.2001
Saha M, Sarkar S, Sarkar B, Sharma BK, Bhattacharjee S, Tribedi P (2016) Microbial siderophores and their potential applications: a review. Envirol Sci Pollut. https://doi.org/10.1007/s11356-015-4294-0
Sanches MS, da Silva CR, Silva LC, Montini VH, Barboza MGL, Guidone GHM, de Oliva BHD, Nishio KN, Galhardi LCF, Vespero EC, Nogueira MCL, Rocha SPD (2021) Proteus mirabilis from community-acquired urinary tract infections (UTI-CA) shares genetic similarity and virulence factors with isolates from chicken, beef and pork meat. Microb Pathog. https://doi.org/10.1016/j.micpath.2021.105098
Sato N, Kawamura K, Nakane K, Wachino JI, Arakawa Y (2013) First detection of fosfomycin resistance gene fosA3 in CTX-M-producing Escherichia coli isolates from healthy individuals in Japan. Microbial Drug Resist. https://doi.org/10.1089/mdr.2013.0061
Skaar EP (2010) The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. https://doi.org/10.1371/journal.ppat.1000949
Stickler DJ (2008) Bacterial biofilms in patients with indwelling urinary catheters. Nat Clin Pract Urol 5:598–608. https://doi.org/10.1038/ncpuro1231
Stickler DJ (2014) Clinical complications of urinary catheters caused by crystalline biofilms: something needs to be done. J Intern Med 276:120–129. https://doi.org/10.1111/joim.12220
Stuck AK, Täuber MG, Schabel M, Lehmann T, Suter H, Mühlemann K (2012) Determinants of quinolone versus trimethoprim-sulfamethoxazole use for outpatient urinary tract infection. Antimicrob Agents Chemother. https://doi.org/10.1128/AAC.05321-11
Tarlton NJ, Moritz C, Adams-Sapper S, Riley LW (2019) Genotypic analysis of uropathogenic Escherichia coli to understand factors that impact the prevalence of β-lactam-resistant urinary tract infections in a community. J Global Antimicrob Resist. https://doi.org/10.1016/j.jgar.2019.03.002
Tolulope A, Ewaoche IS, Ibemologi A (2021) UreC and ZapA virulence genes amplification in clinical specimen of Proteus mirabilis in Bayelsa state, Nigeria. J Microbiol Exp. https://doi.org/10.15406/jmen.2021.09.00317
Valcek A, Roer L, Overballe-Petersen S, Hansen F, Bortolaia V, Leekitcharoenphon P, Hammerum AM (2019) IncI1 ST3 and IncI1 ST7 plasmids from CTX-M-1-producing Escherichia coli obtained from patients with bloodstream infections are closely related to plasmids from E. coli of animal origin. J Antimicrob Chemother. https://doi.org/10.1093/jac/dkz199
Verderosa AD, Totsika M, Fairfull-Smith KE (2019) Bacterial biofilm eradication agents: a current review. Front Chem. https://doi.org/10.3389/fchem.2019.00824
Versalovic J, Koeuth T, Lupski R (1991) Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial enomes. Nucleic Acids Res. https://doi.org/10.1093/nar/19.24.6823
Vieira DC, Lima WG, de Paiva MC (2020) Plasmid-mediated quinolone resistance (PMQR) among Enterobacteriales in Latin America: a systematic review. Mol Biol Rep. https://doi.org/10.1007/s11033-019-05220-9
Walker KE, Moghaddame-Jafari S, Lockatell CV, Johnson D, Belas R (1999) ZapA, the IgA-degrading metalloprotease of Proteus mirabilis, is a virulence factor expressed specifically in swarmer cells. Mol Microbiol. https://doi.org/10.1046/j.1365-2958.1999.01401.x
Wang M, Guo Q, Xu X, Wang X, Ye X, Wu S, Wang M (2009) New plasmid-mediated quinolone resistance gene, qnrC, found in a clinical isolate of Proteus mirabilis. Antimicrob Agents Chemother. https://doi.org/10.1128/AAC.01400-08
Wilson DN, Hauryliuk V, Atkinson GC, O’Neill AJ (2020) Target protection as a key antibiotic resistance mechanism. Nat Rev Microbiol 18:637–648. https://doi.org/10.1038/s41579-020-0386-z
Wirtz VJ, Dreser A, Gonzales R (2010) Trends in antibiotic utilization in eight Latin American countries, 1997-2007. Rev Panam Salud Publ. https://doi.org/10.1590/s1020-49892010000300009
Wollheim C, Guerra IMF, Conte VD, Hoffman SP, Schreiner FJ, Delamare APL, da Costa SOP (2011) Nosocomial and community infections due to class A extended-spectrum β-lactamase (ESBLA)-producing Escherichia coli and Klebsiella spp. in southern Brazil. Braz J Infect Dis 15(2):138–143. https://doi.org/10.1016/S1413-8670(11)70159-3
Woodford N, Fagan EJ, Ellington MJ (2006) Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum (beta)-lactamases. J Antimicrob Chemother 57:154–155. https://doi.org/10.1093/jac/dki412
Yaiche M, Rafraf I, Guo Q, Mastouri M, Aouni M, Wang M (2014) Type II and type IV topoisomerase mutations in clinical isolates of Morganella morganii harbouring the qnrD gene. Annals Clinl Microb Antimicrob. https://doi.org/10.1186/s12941-014-0034-4
Yamanaka T, Funakoshi H, Kinoshita K, Iwashita C, Horikoshi Y (2020) CTX-M group gene distribution of extended spectrum beta-lactamase-producing Enterobacteriaceae at a Japanese Children’s hospital. J Infect Chemother. https://doi.org/10.1016/j.jiac.2020.05.017
Yu J, Kaper JB (1992) Cloning and characterization of the eae gene of enterohaemorrhagic Escherichia coli O157: H7. Mol Microbiol. https://doi.org/10.1111/j.1365-2958.1992.tb01484.x
Acknowledgements
We thank the State University of Londrina and to the Laboratory of Virology of the State University of Londrina for the supply of the cell lines used in this study.
Availability of data and materials
Not applicable.
Funding
This study was financed by the Coordination for the Improvement of Higher Education Personnel - Brazil (CAPES) Finance Code 001.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Luana Carvalho Silva, Matheus Silva Sanches, Gustavo Henrique Migliorini Guidone, Victor Hugo Montini, Bruno Henrique Dias de Oliva, Arthur Bossi do Nascimento, and Lígia Carla Faccin Galhardi. The first draft of the manuscript was written by Renata Katsuko, Takayama Kobayashi, Eliana Carolina Vespero, and Sergio Paulo Dejato Rocha, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of University State of Londrina (CEP-UEL 1.590.120).
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Silva, L.C., Sanches, M.S., Guidone, G.H.M. et al. Clonal relationship, virulence genes, and antimicrobial resistance of Morganella morganii isolated from community-acquired infections and hospitalized patients: a neglected opportunistic pathogen. Int Microbiol 27, 411–422 (2024). https://doi.org/10.1007/s10123-023-00400-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10123-023-00400-x