Abstract
Background
Gastric cancer (GC) is a cancer of the gastrointestinal tract that is highly malignant and has poor prognosis. Circular RNAs are a class of nonclassical RNA molecules that have been determined to be involved in GC malignancy in various ways. However, the underlying function and mechanism of circTDRD3 in gastric cancer remain largely unknown.
Methods
We analyzed circTDRD3 expression in databases and verified the findings in GC cell lines and tissue specimens. A series of functional gene overexpression and knockdown assays in vivo and in vitro were carried out to investigate the role of circTDRD3 in proliferation and metastasis. Here, we revealed the role of the miR-891b/ITGA2 axis by analyzing bioinformatics datasets. Furthermore, we performed dual-luciferase, fluorescence in situ hybridization, RNA pull-down, and functional rescue experiments to examine the relationships between circTDRD3 and its interacting molecules. Western blot confirmed the positive regulatory role of circTDRD3 in the AKT signaling pathway. A promoting effect of ATF4 on circTDRD3 was determined through chromatin immunoprecipitation.
Results
CircTDRD3 was significantly overexpressed in GC tissues compared with adjacent benign tissue, and its expression level was positively correlated with tumor volume and lymph node metastasis. CircTDRD3 promoted GC cell proliferation and migration in vitro and in vivo. Mechanistically, circTDRD3 exerted a tumor-promoting effect by regulating the miR-891b/ITGA2 axis and AKT signaling pathway in a positive feedback manner mediated by the transcription factor ATF4.
Conclusions
ATF4-mediated circTDRD3 overexpression modulates the proliferation and metastasis of GC cells through the miR-891b/ITGA2 axis in a positive feedback manner.
Similar content being viewed by others
Introduction
Gastric cancer (GC) ranks as the fifth most diagnosed malignancy and has the fourth highest mortality globally, with over one million new cases in 2020 and approximately 769,000 deaths [1]. Despite advances in GC therapeutic strategies, surgical or endoscopic resection remains the preferred therapy choice for GC [2]. Even so, many advanced GC patients do not have access to surgery or have poor prognoses after surgery. The five-year survival rate of GC is 25% worldwide and 31% in areas with advanced medical care [3]. As such, brand-new biomarkers for the early diagnosis of or targeted therapy for GC urgently need to be identified.
Circular RNAs (circRNAs) are a unique category of single-stranded noncoding RNAs, generated by a covalently closed loop connection between a downstream 3ʹ splice site and an upstream 5′ splice site [4]. CircRNAs mostly originate from pre-mRNA back-splicing, comprising exonic circRNAs, intronic circRNAs, and exon–intron circRNAs [5]. Currently, numerous noncoding RNAs in human cells and body fluids have been enriched and detected, and the emerging function of circRNAs in evolution, heredity, tumorigenesis, etc., has also been gradually revealed [6]. CircRNAs lack a 3′ poly(A) tail and 5′ 7-methylguanosine cap, ensuring their resistance to degradation by deadenylases and exonucleases [7]. The roles of circRNAs in proliferation, drug resistance, and metastasis have also been extensively examined [8,9,10,11]. CircRNA has been verified to function as tumor regulatory factors by sponging to microRNA(miRNA) [12] or binding to RNA-binding proteins [13]. Additionally, circRNA shows potential to encode proteins through internal ribosome entry sites or specific methylation sites [14]. Recently, researchers identified a few circRNAs anomalously expressed in GC that serve as miRNA sponges. CircNRIP1 sponges miR-149-5p and upregulates the AKT/mTOR axis to promote gastric cancer metastasis [15]. CircREPS2 is a tumor suppressor in GC, that dampens proliferation, migration, and invasion. Low expression of circREPS2 results in the hyperactivity of miR-558 leading to enhanced RUNX3/β-catenin signal transduction in GC [16]. However, identified circRNAs account for only a limited fraction of total circRNAs, and it is necessary to study their tumorigenic mechanisms in GC in more detail.
Our study uncovered a novel circRNA in GC termed circTDRD3 (hsa_circ_0003441) that functions as a miR-891b sponge to promote GC cell proliferation and metastasis. We hope that our findings are helpful for the treatment and prognosis of GC.
Materials and methods
Tissues
Human tissues used in this study were all from patients who underwent surgery in Wuxi No.2 People’s Hospital in 2019 (Table S3). None of the patients received preoperative chemoradiotherapy. All patients signed an informed consent in a self-conscious state. This study was approved by the Ethics Committee of The Affiliated Wuxi No.2 People’s Hospital of Nanjing Medical University (approval number/ID: J201802).
Cell lines and culture
Gastric cancer cell lines (HGC27, MGC803, MKN45, MKN28, SGC7901 and AGS) and human normal gastric mucosal epithelial cell line (GES-1) were all purchased from the Cell Center of Shanghai Institutes for Biological Sciences. Cells were cultured in RPMI-1640 medium (HyClone, USA), supplemented with 10% fetal bovine serum (Gibco, USA) and 1% penicillin/streptomycin (Gibco, USA) at 37 °C and 5% carbon dioxide.
Quantitative real‑time PCR (qRT-PCR)
The materials and methods used in qRT-PCR were consistent with those reported previously [17]. The specific primers applied are mainly exhibited in Table S1. GAPDH and U6 were considered as baselines for assessing circRNA, mRNA, and miRNA expression relatively in a 2−ΔΔCT calculation method.
RNase R treatment and actinomycin D treatment
Total RNA was harvested and cocultured with RNase R (3 U/mg) at 37 ℃ for 20 min (Geneseed, China). Cells were incubated with 2 mg/ml actinomycin D (Sigma, USA) or DMSO as a control and extracted time dependently (0, 4, 8, 12 and 24 h).
Fluorescence in situ hybridization (FISH)
4% paraformaldehyde (PFA) was used to immobilize cells and 0.1% Triton X-100 (Sigma, USA) was used for permeabilization, followed by hybridization overnight at 37 °C with specific probes. CircTDRD3 with Fam-label and miR-891b with Cy3-label were synthesized by GenePharma (Shanghai, China). The nuclei were tagged with DAPI. We obtained the images with a Leica system.
Cell counting Kit-8 (CCK-8)
We purchased CCK-8 (Dojindo, Japan) for assessing the proliferation of GC cells with specific expression patterns. Cells were cultured in 96-well plates with 10 μL CCK-8 reagent for two hours. The 450 nm absorbance of 96-well plates was evaluated by an enzyme-labeled instrument (Thermo, USA).
Colony formation
Cells were treated with trypsin into a cell suspension and plated in 6-well plates. 2 ml of 4% PFA was applied for fixation two weeks after incubation, and then crystal violet was applied for staining.
Wound healing assay
Equal-width scratches were constructed by a sterile pipette tip, and then the pictures (0 h) were obtained with an inverted microscope (Olympus, Japan). After 24 h of incubation, we carefully removed cellular debris with PBS and obtained the images (24 h).
Transwell
Transwell assay inserts (Corning, USA) were placed in 24-well plates. We added 200 μL serum-free RPMI-1640 with 20,000 cells in the upper chamber and 600 μL complete medium in the lower chamber. The next day, cells were treated with methanol and observed with crystal violet staining next day.
5-Ethynyl-2′-deoxyuridine (EdU)
An overnight culture of 1000 cells/well was performed in 96-well plates at 37 °C. Afterward, a kit set (RiboBio, China) was applied to the cells, and photographs were taken using an Olympus fluorescence microscope.
Luciferase reporter assay
HEK-293T cells were transfected with pmirGLO wild-type or mutant circTDRD3 luciferase plasmids (GenePharma, China) by Lipofectamine 3000 (Invitrogen, USA). Luciferase reporters were cotransfected with miR-891b mimics or positive controls simultaneously. After 48 h, Firefly and Renilla luciferase were detected using a kit from Promega (USA).
RNA pull-down
The biotin-labeled probes were all manufactured by GenePharma (Shanghai, China). The collected RNA was incubated with specific or oligo probes for 2 h at 25 °C. Then, treated RNA was mixed with magnetic beads at 4 °C for the whole night. Finally, RNA mixtures were washed with wash buffer and extracted for qRT‒PCR analysis.
Western blot
The operational details of the western blot were as previously reported [17]. Table S2 contained a list of antibodies applied during this study.
Chromatin immunoprecipitation (ChIP)
1% formaldehyde was adopted to crosslink the target proteins with DNA/RNA, which was then terminated by glycine. Cells were lysed and sonicated on ice before being mixed with ChIP-Grade Protein A/G Magnetic Beads (Thermo, USA) and Activating Transcription Factor 4(ATF4)/IgG antibody overnight at 4 °C with rotation. A small mixture was extracted for western blot to examine the specificity and sensitivity of magnetic beads and antibodies. Finally, DNA extracts and purifications were assessed by qRT‒PCR.
Animal experiment
All animal experiments were approved by the Ethics Committee of Wuxi No.2 People’s Hospital. Five-week-old BALB/c nude mice were injected with transfected cells (1 × 106/100 μL) into the subcutis of the axilla. We measured the volumes of subcutaneous tumors weekly. We harvested the xenograft tumors 4 weeks later and weighed them. Subsequently, these tumors were subjected to immunohistochemistry staining.
Luciferase-labeled transfected cells (1 × 106/100 μL) for metastasis assay in vivo, were injected into each mouse through the caudal vein. In five weeks, bioluminescent imaging was captured by in vivo imaging system (Caliper Life Sciences, USA). The Lung metastases were obtained for HE staining.
Statistical analysis
Each experiment was performed in triplicate and independently. Statistical analysis of experimental data was done with SPSS 20.0 (IBM, USA). The results are presented as the mean ± standard deviation (S.D.). Statistical significance was considered when P < 0.05.
Results
Identification of circRNA expression patterns and the expression of circTDRD3 in GC
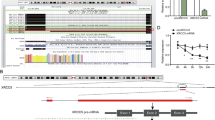
We selected four significantly upregulated circRNAs (P < 0.01, fold change > 100) of GC from a published RNA sequencing data [18], which performed RNA sequence on 15 pairs of gastric cancer tissues. Overall, we verified by qRT‒PCR analysis that circTDRD3 was significantly overexpressed in GC tissues (P < 0.01) (Fig. 1A). CircTDRD3 is a head-to-tail single-stranded 454 nt transcript, formed from exons 2–5 (chr13:61,013,821–61,041,513) of TDRD3 gene (Fig. 1B). Next, Sanger sequencing with RT-PCR product of circTDRD3 verified its head-to-tail splicing (Fig. 1C). qRT-PCR assays also indicated that circTDRD3 was notably upregulated in GC cell lines (Fig. 1D). MKN45 and HGC27 were chosen for follow-up assays due to their high expression levels. Specific convergent and divergent primers were used for complementary DNA (cDNA) and genomic DNA (gDNA) extraction. Subsequently, agarose gel electrophoresis revealed that circTDRD3 could only be amplified by divergent primers from cDNA rather than gDNA (Fig. 1E). Furthermore, we evaluated the attenuation rate of circTDRD3 and TDRD3 in cells added with actinomycin D and the results showed that circTDRD3 contained a stable loop structure instead of linear TDRD3 mRNA (Fig. 1F). Moreover, circTDRD3 showed greater resistance to RNase R degradation than linear TDRD3 (Fig. 1G). qRT‒PCR of RNA isolated from the cytoplasm and nucleus of MKN45 and HGC27 cells revealed that circTDRD3 exhibited a relatively higher RNA level in the cytoplasm than in the nucleus (Fig. 1H), which was consistent with the FISH inferences (Fig. 1I). A 3-year survival analysis was performed on 60 patients, which showed that patients with high circTDRD3 expression indicated a poor prognosis (Fig. 1J).
Identification of circRNA expression patterns and the expression of circTDRD3 in GC. A The four selected circRNAs relative expression was identified in GC tissues with qRT-PCR. B The exonic information of circTDRD3 (circBase ID: hsa_circ_0003441) is illustrated as indicated. C Sanger sequencing validated of back-splicing sites of circTDRD3. D Expression of circTDRD3 (normalized by GAPDH) in GC cell lines compared with the human normal gastric epithelial cell line GES-1. E Verification of specific primers for circTDRD3 by agarose gel electrophoresis. F–G The effect of actinomycin D and RNase R on the relative RNA levels of circTDRD3 and TDRD3 in MKN45 and HGC27 cells was examined by qRT-PCR. H Relative RNA levels of circTDRD3 and TDRD3 mRNA in nuclei (relative to U6) and cytoplasm (relative to GAPDH) fractions. I The subcellular localization of circTDRD3 gene was analyzed by FISH in MKN45 and HGC27 cells. Red indicates circTDRD3. Nuclei were stained with DAPI. Scale bar, 50 μm. J The 3-year survival statistics of 60 patients. Graph represents mean ± SD; *p < 0.05, **p < 0.01, and ***p < 0.001
Given the high expression of circTDRD3 in 60 paired GC tissues, we considered circTDRD3 to be another dependable biomarker for early diagnoses and prognostic assessment. Analysis of the correlation of circTDRD3 expression with clinicopathological features showed that circTDRD3 expression was positively correlated with tumor size and lymph node metastasis (Table S3).
CircTDRD3 promoted GC cell proliferation and migration in vitro
First, two specific small interfering RNAs (siRNAs) targeting circTDRD3 and an overexpression plasmid were designed and their efficiency was measured in HGC27 and MKN45 with qRT‒PCR (Figure S1A-B). Then, we performed colony formation, CCK-8 and EdU assays to explore the role of circTDRD3 on GC cell proliferation. Compared to the control group, GC cell proliferation was significantly reduced when transfected with siRNA (Fig. 2A–C, statistics in Fig S1C–D). The increased circTDRD3 expression notably promoted proliferation after being transfected with the overexpression vector (Fig. 2A–C, statistics in Fig S1C–D). We also analyzed cell cycle proportions via flow cytometry, showing that S-phase percentage was markedly decreased in circTDRD3-knockdown cells. When circTDRD3 was overexpressed, the number of GC cells in S phase increased accordingly, and the proportion of cells in the GO–G1 phases showed a declining tendency (Fig. 2D–E).
CircTDRD3 promoted GC cell proliferation and migration in vitro. A–C MKN45 and HGC27 cells were transfected with control, circTDRD3 siRNA, vector or circTDRD3 plasmid. CCK-8, Colony formation assay and EdU assays were carried out to assess the effect of circTDRD3 on the proliferation of GC cells. Scale bar: 100 μm. D–E The function of circTDRD3 in regulating cell cycle distribution of MKN45 and HGC27 cells was detected by flow cytometry. F Using Transwell assay to explore the effect of circTDRD3 on the cell migration of GC cells. Scale bar: 100 μm. G Western Blot was aimed to detect the cell migration marker proteins in the EMT pathway with the change of circTDRD3 RNA level. Graph represents mean ± SD; *p < 0.05, **p < 0.01, and ***p < 0.001
Analysis of migration ability revealed that circTDRD3 promoted the migration and invasion of GC cells in transwell and wound healing assays, while knockdown of circTDRD3 reduced these abilities (Fig. 2F, statistics in Fig S1E–F). In addition, western blot assay (Fig. 2G) and its statistical analysis (Fig S5 A–C) exhibited that expression of cell migration markers in the EMT pathway was markedly elevated in cells overexpressing circTDRD3 and decreased in cells with circTDRD3 knockdown.
Comparatively, the results of overexpression experiments with circTDRD3 low expression cell line SGC7901 were in favor of a significantly enhanced ability of proliferation and migration (Fig S3).
In summary, all these findings indicated that circTDRD3 expression was positively related to the migration and proliferation of GC cell.
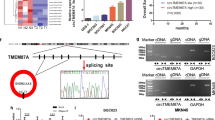
CircTDRD3 functioned as a miR-891b sponge
CircRNAs usually exert their biological function by sponging specific miRNAs [12]. To determine the target miRNAs of circTDRD3, we studied miRanda (https://microrna.org/microrna.home.do), RNAhybird (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/) and CircInteractome (https://circinteractome.nia.nih.gov/index.html) and identified four candidate miRNAs (Fig. 3A). To pull down circTDRD3 in GC cells, we first employed a specific biotin-labeled circTDRD3 probe. qRT‒PCR assays demonstrated that the amount of circTDRD3 pulled down by the specially designed probe was much higher than that pulled down by the oligo probe (Fig. 3B). Next, we performed RNA pull-down experiments and found miR-891b highest expression among the four candidate miRNAs in both MKN45 and HGC27 cells (Fig. 3C). The predicted binding sites between miR-891b and circTDRD3 are exhibited in Fig. 3D and Luc-circTDRD3-wt luciferase activity was significantly decreased by miR-891b overexpression. (Fig. 3E). By contrast, the luciferase activity was not different in the Luc-circTDRD3-mut group. We also designed biotin-labeled wt- and mutant- miR-891b constructs to verify the interactions between miR-891b and circTDRD3. Bio-miR-891b-wt bound to more circTDRD3 than Bio-miR-891b-mut (Fig. 3F).
CircTDRD3 functioned as a miR-891b sponge. A The overlap of the Venn diagram showed that there are four candidate targeted miRNAs of circTDRD3 predicted by miRanda, RNAhybrid and CircInteractome. B We confirm the enrichment of circTDRD3-specific probe relative to oligo probe by qRT-PCR. C The four candidate miRNAs were pulled down by a biotin-labeled circTDRD3-specific probe in MKN45 and HGC27 cells and the expression levels of pull-down product were measured by qRT-PCR. D The predicted binding sites of miR-891b to circTDRD3. E Dual luciferase report assay demonstrated that miR-891b and circTDRD3 bind to each other via the predicted region. F Pull down with wild-type or mutant miR-891b in MKN45 and HGC27 cells and detect the RNA levels of circTDRD3. G The FISH assay revealed the colocalization of circTDRD3 and miR-891b in MKN45 and HGC27 cell cytoplasm. Scare bar = 50 μm. H Western blot analysis was performed to investigate the protein levels of E-cadherin, N-cadherin and Vimentin when the expression pattern of miR-891b changed and the restorative effect of miR-891b on circTDRD3 function in EMT pathway. Graph represents mean ± SD; *p < 0.05, **p < 0.01, and ***p < 0.001
In addition, RNA FISH experiments demonstrated that miR-891b and circTDRD3 shared a common subcellular localization in MKN45 and HGC27 cells (Fig. 3G). Western blot (Fig. 3H) and its statistical analysis (Fig S5D–I) revealed that the miR-891b mimics and anti-miR-891b reversed the effect of circTDRD3 on the EMT pathway.
ITGA2 was identified as a downstream target of miR-891b
We used the online databases miRDB (http://www.mirdb.org/), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/index.html/) and TargetScan (http://www.targetscan.org/vert_71/) to identify thirteen candidate genes as targets of miR-891b (Fig. 4A). Integrin, alpha 2 (ITGA2) was reported to promote GC progression and was significantly upregulated in TCGA samples accessed via the UALCAN database (http://ualcan.path.uab.edu/) (Fig. 4B). Moreover, the Kaplan‒Meier Plotter database (https://kmplot.com/analysis/) indicated that high expression of ITGA2 in TCGA samples might indicate a poor prognosis (Fig. 4C). ITGA2 expression was detected in all 60 paired GC tissues (Fig. 4D), confirming TCGA results.
ITGA2 was identified as a downstream target of miR-891b. A The Venn diagram showed that there are thirteen downstream target genes of miR-891b by exploring miRDB, miRTarBase and Targetscan. B The expression level of ITGA2 in TCGA samples accessed via the UALCAN database. C The Kaplan–Meier Plotter database exhibited the association between the levels of ITGA2 and prognoses in TCGA samples. D The expression levels of ITGA2 were validated in 60 paired GC tissues. E–F The levels of ITGA2 mRNA and protein were detected by qRT-PCR and western blot in MKN45 and HGC27 cells, and there was a regulatory relationship between miR-891b and ITGA2. G Western blot assay detected the protein levels of ITGA2 when we overexpressed circTDRD3 or knocked down miR-891b. H Dual luciferase report assay confirmed the combination between miR-891b and ITGA2. Graph represents mean ± SD; *p < 0.05, **p < 0.01, and ***p < 0.001
We undertook qRT-PCR and western blot for detecting the effect that miRNA inhibitors or mimics had on ITGA2 expression and found that ITGA2 expression was upregulated with miR-891b knockdown. ITGA2 was also expressed at a lower level when miR-891b was overexpressed. (Fig. 4E–F, statistics in Fig S5J–K). Our hypothesis that the protein level of ITGA2 could be rescued by a miR-891b inhibitor in circTDRD3-overexpressing cells and repressed by miR-891b mimics in circTDRD3-downregulated cells was confirmed by western blot (Fig. 4G, statistics Fig S5 L-M). In addition, miR-891b mimics decreased wild-type ITGA2 luciferase reporter activity and caused no significant changes in mutant ITGA2 (Fig. 4H).
CircTDRD3 promoted cell proliferation, migration and invasion through upregulating ITGA2 in GC
From the above experimental results, we concluded that circTDRD3 promoted GC malignant progression by sponging miR-891b, thereby amplifying the tumorigenic effect of ITGA2. Thus, we performed rescue experiments to verify the circTDRD3/miR-891b/ITGA2 competing endogenous RNA (ceRNA) regulatory network. We suppressed the expression of ITGA2 in circTDRD3-overexpressed MKN45 and HGC27 cell and increased the level of ITGA2 in circTDRD3-knockdown cells. Colony formation, EdU, and cell cycle analyses revealed that inhibition of ITGA2 weakened the proliferation of cells overexpressing circTDRD3 and that a high level of ITGA2 rescued the impaired proliferation ability of circTDRD3-knockdown cells (Fig. 5A–F). In line with these results, transwell and wound healing assays demonstrated that the reduction in migration and invasion ability caused by circTDRD3-knockdown cells was strengthened by increased levels of ITGA2 (Fig S2A–D).
CircTDRD3 promoted cell proliferation, migration and invasion through upregulating ITGA2 in GC. A–B Colony formation assay of the cell proliferation in MKN45 and HGC27 cells. C–D EdU staining in revealing the cell proliferation in MKN45 and HGC27 cells. Scale bar: 100 μm. E–F Cell cycle analysis indicated the cell proliferation ability in MKN45 and HGC27 cells. Graph represents mean ± SD; *p < 0.05, **p < 0.01, and ***p < 0.001
This section illustrated that circTDRD3 promoted proliferation, migration and invasion through upregulating ITGA2 in GC cells.
CircTDRD3 promoted GC cell growth and lung metastasis by upregulating ITGA2 in vivo
We attempted to determine the regulatory function of circTDRD3 in vivo and constructed stable sh-circTDRD3 and ov-circTDRD3 cells with or without stable knockdown or overexpression of ITGA2. First, we generated xenograft tumors in nude mice and measured their weights and volumes weekly. The tumors derived from circTDRD3-overexpressing cells grew faster, while those generated from circTDRD3-knockdown cells exhibited smaller volumes and lighter weights (Fig. 6A–C). Furthermore, upregulation of ITGA2 intensified the positive effects of circTDRD3 on tumor growth, and depleting ITGA2 reversed these effects (Fig. 6A–C). Subsequently, the xenograft tumors were gathered and processed for Ki67 staining to detect proliferation. The IHC scores of Ki67 staining in xenograft tumor tissues were in line with the above results (Fig. 6D–E). Next, we constructed lung metastasis models by injecting the indicated GC cells through the caudal vein to examine whether circTDRD3 promotes GC cell metastasis in vivo. Five weeks after injection, we checked for the presence of metastases in the lungs and the size of metastases through an IVIS (Fig. 6F–G). Finally, HE staining was performed on lung tissues after they had been harvested. (Fig. 6H–I).
CircTDRD3 prompted GC cell growth and lung metastasis by upregulating ITGA2 in vivo. A–C Generated xenograft tumors in nude mice and measured the weights and volumes of subcutaneous tumors every week. D–E IHC staining and scores of ki67 after gathering the xenograft tumors. Scale bar: 100 μm. F–G The size and quantification of metastases in the lungs were observed through IVIS. H–I Representative images and quantification of HE staining of lung metastases. Scale bar: 1 mm. Graph represents mean ± SD; *p < 0.05, **p < 0.01, and ***p < 0.001
Taken together, the above results suggested that circTDRD3 enhanced GC cell growth and metastasis by upregulating ITGA2 in vivo.
CircTDRD3/miR-891b/ITGA2 promoted GC tumorigenesis by regulating the AKT signaling pathway in a positive feedback manner mediated by ATF4
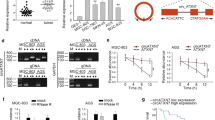
By studying KEGG (https://www.kegg.jpitga/) database, we noticed that ITGA2 was one of the critical initiators of the AKT pathway [19]. Considering that western blotting results implied that overexpressed circTDRD3 could induce the activation of AKT signal pathway (Fig S4 A–E), we hypothesized that circTDRD3 could motivate the AKT pathway via circTDRD3/miR-891b/ITGA2 axis in GC.
To test our hypothesis, we examined the expression levels of key proteins in the AKT pathway using western blotting. In addition to stable knockdown and overexpression of ITGA2, we also used the pathway inhibitor MK2206 and the agonist SC79. First, we found that differential ITGA2 expression did not significantly influence total AKT protein levels, while phosphorylated AKT (p-AKT) correlated significantly with ITGA2 (Fig. 7A, statistics in Fig S6). In addition, we detected proteins involved in AKT signaling and found that the circTDRD3/miR-891b/ITGA2 axis plays a catalytic role in activating Foxo1 via phosphorylation (p-Foxo1) and upregulating the expression of ATF4 (Fig. 7A). Cyclin D1 was upregulated by ITGA2 overexpression and downregulated by ITGA2 knockdown. Similarly, ITGA2 exerted a positive regulatory effect on cyclin D1 in cells treated with the agonist or the inhibitor (Fig. 7A).
CircTDRD3/miR-891b/ITGA2 promoted GC tumorigenesis by regulating the AKT signaling pathway in a positive feedback manner mediated by ATF4. A Western blot was utilized to evaluate the levels of key protein molecules (ITGA2, p-AKT, AKT, ATF4, p-Foxo1, cyclin D1) of the AKT signaling pathway in MKN45 and HGC27 cells. B The complete binding sequences between ATF4 and TDRD3 were verified by the JASPAR website. C Schematic illustration of the design of primers specific for TDRD3, pre-TDRD3 and circTDRD3. D qRT-PCR was performed to assess the effect of si-ATF4 and ATF4 on the RNA levels of TDRD3, pre-TDRD3 and circTDRD3. D Luciferase assay showed enhanced luciferase reporter activity after transfection of ATF4 plasmid. F ChIP-PCR assay and subsequent agarose gel electrophoresis confirmed the mutual combination of TDRD3 promoter and ATF4. H Graphical abstract of the mechanism of circTDRD3 cancer-promoting effect in GC cells. Graph represents mean ± SD; *p < 0.05, **p < 0.01, and ***p < 0.001
Additionally, we found that ATF4, an important transcriptional regulator [20,21,22], was also significantly elevated in GC. Therefore, we wondered whether ATF4 could promote the circularization of TDRD3 into circTDRD3 transcriptionally. First, we verified that ATF4 and TDRD3 had complimentary binding sequences through JASPAR (https://jaspar.genereg.net/) (Fig. 7B). Second, different primers were specifically designed to detect the posttranscriptional levels of TDRD3, pre-TDRD3 and circTDRD3 (Fig. 7C). We found that elevated expression of ATF4 increased the level of circTDRD3, which correspondingly decreased after ATF4 knockdown (Fig. 7D). ATF4 also had the same effect on linear TDRD3 and pre-TDRD3 mRNA levels (Fig. 7D). Third, as shown by the luciferase reporter assay, ATF4 activated the TDRD3 promoter region (Fig. 7E). Finally, we selected an ATF4-specific antibody to perform ChIP‒PCR assays and subsequent agarose gel electrophoresis and found that ATF4 could closely bind to the predicted promoter region of TDRD3 (Fig. 7F–G).
Discussion
Recently, circRNAs have been widely found to be abnormally expressed in atherosclerosis [23], Alzheimer's disease [24], leukemia [25], malignant tumors [26] and other diseases. CircRNA cannot be easily degraded by exonuclease, so they have advantages in clinical diagnoses, treatment, or prognostication of various diseases. Therefore, we focused on expression patterns and mechanisms of circRNA in GC.
In this study, circTDRD3 originating from TDRD3 was identified to be significantly overexpressed in GC cells through database analysis and screening. First, we determined the circularized structure based on gene sequence, expression levels, cellular sublocalization and stability analyses. Then, to illustrate the function of circTDRD3 in the malignant transformation of GC, we carried out a series of functional experiments. Clinicopathological feature analysis showed that GC tumors with high circTDRD3 expression exhibited higher levels of tumor growth and lymph node metastasis. After modifying the circTDRD3 expression patterns, we found that circTDRD3 promoted GC cell invasion, migration and proliferation in vitro and in vivo.
Next, we focused our efforts on exploring the mechanism of circTDRD3 in GC progression and development. Many studies have reported circRNAs related to GC [27]. The main mechanism of circRNAs is functioning as a sponge to regulate miRNAs [28]. In our study, we identified four candidate miRNAs via the miRanda, RNAhybrid and CircInteractome databases. By verifying the interaction between circTDRD3 and the candidate miRNAs, miR-891b was identified as a downstream target of circTDRD3. Similarly, we identified ITGA2 as downstream of the circTDRD3/miR-891b regulatory axis by analyzing the interaction data from miRDB, TargetScan and miRTarBase and related data from TCGA.
Phosphorylated AKT activates its downstream molecules and participates in cell survival, growth, metabolism, tumorigenesis and metastasis [29]. Aberrant hyperphosphorylation of AKT can lead to many cancers, thus increasing the development of AKT-targeted inhibitors to improve the clinical outcome of advanced tumors [30]. Compared with AKT-targeting monotherapy, EGFR or CDK inhibitors combined with these agents have performed better in preclinical studies [31,32,33]. However, for the effectiveness of targeted drugs, the types of drugs, doses of different inhibitor combinations, poor tolerability and serious toxic effects and side effects need to be taken into consideration [34]. Therefore, therapies targeting the AKT pathway need to be improved to overcome the existing problems. According to the KEGG database, ITGA2 is an initiator of the AKT pathway [35]. We found that overexpressed circTDRD3 stimulates the AKT pathway via the circTDRD3/miR-891b/ITGA2 regulatory axis in GC, thereby activating AKT pathway molecules, including Cyclin D1, Foxo1 and ATF4.
ATF4 has aberrant expression patterns in a variety of tumors [36]. When tumor cells are undergoing a stress response, the expression level of ATF4 can be significantly increased to promote cancer progression [37], such as targeting the glutamate antiporter xCT/SCL7A11 and increasing the transcription of xCT [38]. Here, we sought to determine the regulatory role of ATF4 in the circularization of circTDRD3 and found that ATF4 can combine with the predicted region of TDRD3 promoter and promote circularization of circTDRD3. This finding increases our understanding of the circRNA regulatory network in GC.
However, our research was abundant in vitro studies, but did not initiate profounder animal experiments, such as knockout mice, or clinical trials, which means that the future in vivo application of the circTDRD3 is unclear, suggesting that further exploration is needed in the future.
Conclusion
Collectively (Fig. 7H), we reported a novel circRNA that is significantly overexpressed in GC, termed circTDRD3, enhancing GC cell proliferation and metastasis. We inventively propose a positive feedback loop between circTDRD3 and AKT signaling that may bring new hope to improve the outcome of precision cancer treatment by interfering with this loop signaling.
Data availability
The datasets in this study are available from the corresponding author on reasonable request.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J Clin. 2021;71(3):209–49.
Smyth EC, Nilsson M, Grabsch HI, van Grieken NCT, Lordick F. Gastric cancer. Lancet. 2020;396(10251):635–48.
Sexton RE, Al Hallak MN, Diab M, Azmi AS. Gastric cancer: a comprehensive review of current and future treatment strategies. Cancer Metastasis Rev. 2020;39(4):1179–203.
Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17(11):679–92.
Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–42.
Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32(5):309–16.
Guo Y, Wei X, Peng Y. Structure-mediated degradation of CircRNAs. Trends Cell Biol. 2020;30(7):501–3.
Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–81.
Liu F, Zhang H, Xie F, Tao D, Xiao X, Huang C, Wang M, Gu C, Zhang X, Jiang G. Hsa_circ_0001361 promotes bladder cancer invasion and metastasis through miR-491-5p/MMP9 axis. Oncogene. 2020;39(8):1696–709.
Xu J, Ji L, Liang Y, Wan Z, Zheng W, Song X, Gorshkov K, Sun Q, Lin H, Zheng X, et al. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct Target Ther. 2020;5(1):298.
Zeng K, He B, Yang BB, Xu T, Chen X, Xu M, Liu X, Sun H, Pan Y, Wang S. The pro-metastasis effect of circANKS1B in breast cancer. Mol Cancer. 2018;17(1):160.
Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, Ren D, Ye X, Li C, Wang Y, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79.
Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10(8):3503–17.
Wang J, Zhu S, Meng N, He Y, Lu R, Yan GR. ncRNA-encoded peptides or proteins and cancer. Mol Ther. 2019;27(10):1718–25.
Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, Xu P, Sun G, Xu J, Lv J, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18(1):20.
Guo X, Dai X, Liu J, Cheng A, Qin C, Wang Z. Circular RNA circREPS2 acts as a sponge of miR-558 to suppress gastric cancer progression by regulating RUNX3/beta-catenin signaling. Mol Ther Nucleic Acids. 2020;21:577–91.
Wang N, Lu K, Qu H, Wang H, Chen Y, Shan T, Ge X, Wei Y, Zhou P, Xia J. CircRBM33 regulates IL-6 to promote gastric cancer progression through targeting miR-149. Biomed Pharmacother. 2020;125:109876.
Shen Y, Zhang J, Fu Z, Zhang B, Chen M, Ling X, Zou X. Gene microarray analysis of the circular RNAs expression profile in human gastric cancer. Oncol Lett. 2018;15(6):9965–72.
Huang W, Zhu J, Shi H, Wu Q, Zhang C. ITGA2 overexpression promotes esophageal squamous cell carcinoma aggression via FAK/AKT signaling pathway. Onco Targets Ther. 2021;14:3583–96.
Huang P, Peslak SA, Lan X, Khandros E, Yano JA, Sharma M, Keller CA, Giardine B, Qin K, Abdulmalik O, et al. The HRI-regulated transcription factor ATF4 activates BCL11A transcription to silence fetal hemoglobin expression. Blood. 2020;135(24):2121–32.
Kitakaze K, Oyadomari M, Zhang J, Hamada Y, Takenouchi Y, Tsuboi K, Inagaki M, Tachikawa M, Fujitani Y, Okamoto Y, et al. ATF4-mediated transcriptional regulation protects against beta-cell loss during endoplasmic reticulum stress in a mouse model. Mol Metab. 2021;54: 101338.
Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J, Yuan CL, Krokowski D, Wang S, Hatzoglou M, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol. 2013;15(5):481–90.
Cao Q, Guo Z, Du S, Ling H, Song C. Circular RNAs in the pathogenesis of atherosclerosis. Life Sci. 2020;255: 117837.
Huang JL, Su M, Wu DP. Functional roles of circular RNAs in Alzheimer’s disease. Ageing Res Rev. 2020;60: 101058.
Wu Z, Sun H, Li J, Jin H. Circular RNAs in leukemia. Aging. 2019;11(13):4757–71.
Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–65.
Li R, Jiang J, Shi H, Qian H, Zhang X, Xu W. CircRNA: a rising star in gastric cancer. Cell Mol Life Sci: CMLS. 2020;77(9):1661–80.
Cheng Z, Yu C, Cui S, Wang H, Jin H, Wang C, Li B, Qin M, Yang C, He J, et al. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat Commun. 2019;10(1):3200.
Revathidevi S, Munirajan AK. Akt in cancer: mediator and more. Semin Cancer Biol. 2019;59:80–91.
Uko NE, Guner OF, Matesic DF, Bowen JP. Akt pathway inhibitors. Curr Top Med Chem. 2020;20(10):883–900.
Viswanathan A, Kute D, Musa A, Konda Mani S, Sipila V, Emmert-Streib F, Zubkov FI, Gurbanov AV, Yli-Harja O, Kandhavelu M. 2-(2-(2,4-dioxopentan-3-ylidene)hydrazineyl)benzonitrile as novel inhibitor of receptor tyrosine kinase and PI3K/AKT/mTOR signaling pathway in glioblastoma. Eur J Med Chem. 2019;166:291–303.
Cairns J, Fridley BL, Jenkins GD, Zhuang Y, Yu J, Wang L. Differential roles of ERRFI1 in EGFR and AKT pathway regulation affect cancer proliferation. EMBO Rep. 2018;19(3):e44767.
Lai IC, Lai GM, Chow JM, Lee HL, Yeh CF, Li CH, Yan JL, Chuang SE, Whang-Peng J, Bai KJ, et al. Active fraction (HS7) from Taiwanofungus camphoratus inhibits AKT-mTOR, ERK and STAT3 pathways and induces CDK inhibitors in CL1-0 human lung cancer cells. Chin Med. 2017;12:33.
Lacouture M, Sibaud V. Toxic side effects of targeted therapies and immunotherapies affecting the skin, oral mucosa, hair, and nails. Am J Clin Dermatol. 2018;19(Suppl 1):31–9.
Ma L, Sun Y, Li D, Li H, Jin X, Ren D. Overexpressed ITGA2 contributes to paclitaxel resistance by ovarian cancer cells through the activation of the AKT/FoxO1 pathway. Aging. 2020;12(6):5336–51.
Wortel IMN, van der Meer LT, Kilberg MS, van Leeuwen FN. Surviving stress: modulation of ATF4-mediated stress responses in normal and malignant cells. Trends Endocrinol Metab. 2017;28(11):794–806.
Singleton DC, Harris AL. Targeting the ATF4 pathway in cancer therapy. Expert Opin Ther Targets. 2012;16(12):1189–202.
Chen D, Fan Z, Rauh M, Buchfelder M, Eyupoglu IY, Savaskan N. ATF4 promotes angiogenesis and neuronal cell death and confers ferroptosis in a xCT-dependent manner. Oncogene. 2017;36(40):5593–608.
Acknowledgements
This work was supported by Key Project of Scientific Research from Jiangsu Commission of Health (Grant numbers: ZDB2020026); Wuxi Taihu Lake Talent Plan, Team in Medical and Health Profession; Wuxi Medical Key Discipline Construction Project, Medical Development Discipline; Wuxi Science and Technology Development Fund from Wuxi Science and Technology Bureau (Grant numbers: N20201002); Natural Science Foundation of Jiangsu Province Youth Fund (Grant numbers: BK20210065); Youth Fund Project from Wuxi Commission of Health (Grant numbers: Q202013); Precision Medicine Key Project from the Health Commission of Wuxi (Grant numbers: J202109).
Author information
Authors and Affiliations
Contributions
JX and YC designed and instructed this study; PZ, HQ, KS and XC performed most experiments; ZZ and QZ contributed to the animal experiments; ZL and YZ collected and sorted the clinical data; NW and KD conducted the statistical analysis; PZ drafted this manuscript; LW, TS and GF gave revised opinions.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics statement
This study was approved by the ethics committee of Wuxi No.2 People’s Hospital (approval number/ID: J201802).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhou, P., Qu, H., Shi, K. et al. ATF4-mediated circTDRD3 promotes gastric cancer cell proliferation and metastasis by regulating the miR-891b/ITGA2 axis and AKT signaling pathway. Gastric Cancer 26, 565–579 (2023). https://doi.org/10.1007/s10120-023-01392-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-023-01392-3