Abstract
Objective
It remains unknown whether individuals with a family history (FH) of gastric cancer (GC) are associated with aberrant DNA methylation. The aim of this study was to investigate the association between aberrant DNA methylation and FH of GC.
Design
Using quantitative MethyLight assay, MOS, miR124a-3, NKX6-1, EMX1, CDH1, and TWIST1 methylation levels in the noncancerous gastric mucosa was compared between subjects with and without FH based on GC and Helicobacter pylori (Hp) infection. Changes in the methylation levels were evaluated over time after Hp eradication.
Results
In Hp-positive GC patients, MOS (P < 0.001), CDH1 (P < 0.001), and TWIST1 (P = 0.004) methylation were decreased in subjects with FH (n = 64) than in those without FH (n = 58). In Hp-positive controls, MOS methylation was lower in subjects with FH (n = 73) than in those without FH (n = 50) (P = 0.042), while miR124a-3 (P = 0.006), NKX6-1 (P < 0.001), and CDH1 (P < 0.001) methylation were higher in subjects with FH. CDH1 methylation constantly decreased from 2 years in GC patients and 3–4 years in controls after Hp eradication (all P < 0.001). A persistent decrease in methylation over time was not observed in other genes after eradication.
Conclusion
The methylation of MOS and CDH1 provided an association between aberrant DNA methylation and gastric carcinogenesis in FH of GC, a useful marker for GC risk in individuals with FH. Furthermore, CDH1 methylation decreased after Hp eradication.
Similar content being viewed by others
Introduction
Aberrant DNA methylation is a major epigenetic mechanism and is profoundly involved in gastric carcinogenesis [1]. Gene silencing of most tumor suppressor genes and tumor-related genes, mediated by regional hypermethylation in promoter regions, is almost exclusively associated with CpG islands and has been widely studied in gastric cancer (GC) [2]. Additionally, aberrant DNA methylation has been observed in the noncancerous mucosa of patients with GC and non-neoplastic gastric mucosa of subjects without GC, at levels correlating with the risk of cancer development, producing a so-called ‘epigenetic field for cancerization’ [1, 3]. Helicobacter pylori (Hp)-induced chronic inflammation plays a direct role in the induction of aberrant DNA methylation [4,5,6,7], and this methylation may partially reverse in certain genes with the eradication of infection [8, 9]. Conversely, residual methylation in gastric mucosa after Hp eradication might be associated with the risk of developing GC [10].
In GC, family history (FH) is a well-established risk factor [11]. Most GCs are sporadic, with approximately 10% of GCs attributed to familial clustering; however, nearly 1–3% of GCs arise from inherited GC predisposition syndromes, such as hereditary diffuse gastric carcinoma and familial adenomatous polyposis [12]. The risk of GC in individuals with FH is approximately threefold higher than in those without FH [11]. A Korean study has shown that in women with both FH and Hp infection the risk of GC increased by fivefold when compared with those without these predisposing factors [13]. Indeed, family members share various factors, from exposure to the same carcinogens (i.e., nitrogen, cigarette smoke, and alcohol consumption) to similar levels of hygiene, dietary habits (salty and spicy food, smoked food), bacterial virulence, and genetic susceptibility [14]. Single nucleotide polymorphisms related to cytokines such as transforming growth factor-β1 and interleukin-1 have been investigated to establish genetic predisposition in GC patients with FH [14, 15]. Additionally, GC patients with FH present a lower cancer stage (I–II) at diagnosis than those without FH, but overall survival was not different between patients with and without FH [16].
Although associations between GC risk and aberrant DNA methylation have been previously reported, evidence indicating the association of FH of GC with DNA methylation levels in GC patients and healthy individuals remains limited. If FH of GC is associated with DNA methylation levels even in healthy individuals, methylation levels could serve as a potential marker of molecular changes in the multistep process of GC development. In this study, we hypothesized that aberrant DNA methylation of GC-related genes is associated with GC development in subjects with FH. Thus, the aim of this study was to investigate the methylation of these genes in noncancerous gastric mucosa from patients with GC and controls, considering FH of GC and Hp infection. Furthermore, the long-term effects of Hp eradication on epigenetic molecular alterations were investigated in these patients. In this study, six genes, MOS, miR124a-3, NKX6-1, EMX1, CDH1, and TWIST1 were selected. The methylation level of MOS was significantly elevated in patients with metachronous gastric neoplasm (gastric dysplasia and early GC) compared patients without that in our previous researches [17]. In addition, decrease of MOS methylation level following H. pylori eradication was significant among controls without intestinal metaplasia (IM) (P value for slope < 0.05) [18]. miR124a-3, NKX6-1 and EMX1 methylation were an informative marker for predicting the risk of metachronous GC in patients after endoscopic resection in Japanese multicenter prospective cohort study [19]. CDH1 is a tumor suppressor gene and has been reported frequently hypermethylated in GC and also suggested marker of GC development [20, 21]. TWIST1 encodes a basic-helix-loop-helix transcription factor called Twist1 that regulates metastasis and epithelial–mesenchymal transition [22]. Although Twist1 protein may have tumor or metastasis-promoting effects, paradoxically, hypermethylated TWIST1 has been reported in GC and other cancer [22].
Material and methods
Study subjects
The study subjects who had gastrointestinal symptoms such as epigastric discomfort, nausea, vomiting or presented for regular check-up for surveillance of GC at the Seoul National University Bundang Hospital from February 2006 to September 2019 were consecutively enrolled. The subjects with a previous history of Hp eradication or gastrectomy, and past Hp infection were excluded. In total, 328 subjects including 181 healthy controls and 147 patients with GC were selected (Fig. 1). The controls showed no evidence of GC, dysplasia, mucosa-associated lymphoid tissue lymphoma, esophageal cancer, or peptic ulcer. All subjects, who provided informed consent, were asked to complete a questionnaire, requesting data regarding age, sex, smoking and drinking habits, history of Hp eradication, and FH of GC. A “positive family history” was defined as having any first-degree relatives (parent, sibling, or offspring) diagnosed with GC. Hp-positive subjects were treated with a standard 7-day triple therapy (esomeprazole 40 mg b.i.d., clarithromycin 500 mg b.i.d., and amoxicillin 1000 mg b.i.d.) for Hp eradication. To evaluate Hp eradication, 13C-Urea breath testing was performed at least 4 weeks after completion of the eradication therapy. The study protocol was approved by the Ethical Committee at Seoul National University Bundang Hospital (IRB number: B-1012/117-013).
H. pylori testing and histology
During endoscopy, 10 biopsy specimens were obtained to determine the Hp infection status, as well as histological evaluation. Among these specimens, two from the greater curvature side of the antrum and two from the body were fixed in formalin to assess the presence of Hp using modified Giemsa staining, and to examine the degree of inflammatory cell infiltration, atrophy, and IM, determined by hematoxylin and eosin staining. The histologic features of the gastric mucosa were recorded using the updated Sydney scoring system. One specimen from each, the lesser curvature of the antrum and the body, was used for rapid urease testing (CLOtest, Delta West, Bentley, Australia). Subsequently, two specimens from the antrum and the body were used for culture, and organisms present were identified as Hp by Gram staining, colony morphology, and positive oxidase, catalase, and urease reactions. The remaining biopsy specimens and GC tissues were immediately frozen at -70 °C until DNA extraction.
Positivity for CLOtest, Giemsa staining, and culture was defined as current Hp infection. If results obtained from three biopsy-based tests were negative, 13C-urea breath test (UBiTkit; Otsuka Pharmaceutical, Tokyo, Japan) and serum immunoglobulin G antibodies for Hp detected by an enzyme-linked immunosorbent assay (Genedia Hp ELISA; Green Cross Medical Science Corp., Eumseong, South Korea) were performed. Hp bacteria are spontaneously disappeared in severely atrophic mucosa and advanced gastric diseases once they have caused gastric atrophy and IM after long-time colonization, and the spontaneous seroconversion of anti-Hp antibodies often occurs [23]. Therefore, among the patients with negative results from all the above clinical tests and no prior history of Hp eradication, the true negative Hp infection was diagnosed as follows: PG I/II ratio > 3 and no or mild atrophy and IM either in the antrum and body. Positivity for the Hp serology test and/or an Hp eradication history indicated a past Hp infection; individuals with a past Hp infection were excluded from this analysis.
DNA extraction, bisulfite modification, and MethyLight assay
Genomic DNA was extracted directly from noncancerous antral biopsy specimens using sodium bisulfite. Briefly, specimens were homogenized in proteinase K solution [20 mmol/L Tris–HCl (pH 8.0), 10 mmol/L ethylenediaminetetraacetic acid, 0.5% sodium dodecyl sulfate, and 10 mg/mL proteinase K] using a sterile micropestle, followed by incubation for 3 h at 52 °C. DNA was isolated from homogenates using phenol/chloroform extraction and ethanol precipitation. Genomic DNA (1 µg) was bisulfite modified using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA) by following the manufacturer’s instructions. The methylation status of six genes (MOS, miR124a-3, NKX6-1, EMX1, CDH1, and TWIST1) from bisulfite-modified DNA samples was quantified using real-time polymerase chain reaction (PCR)-based MethyLight technology [24]. MethyLight, as a sensitive, high-throughput methylation assay, allows the highly specific detection of methylation using probes that cover methylation sites, as well as methylation-specific primers. The primer and probe sequences used in the reaction are shown in Supplementary Table S1. The quantified level of each gene was reported as a percentage of methylated reference (PMR), which is the relative methylation ratio of the target gene to the ALU gene of a sample, divided by the ratio of the target gene to the ALU gene of sodium bisulfite and CpG methyltransferase (M.SssI)-treated sperm DNA, multiplied by 100 [24].
Statistical analysis
Comparisons of demographic and clinicopathologic variables were performed using the Student t test or Chi-square test (Fisher’ s exact test) for continuous and categorical variables, respectively. The Mann–Whitney U test was used to compare medians when the sample size was less than 30. All statistical analyses were performed using SPSS (version 21.0; SPSS, Chicago, IL, USA). A P value < 0.05 was considered significant.
Results
Subjects characteristics
The clinicopathological characteristics of the study subjects are summarized in Table 1. Overall, 328 patients, including 147 patients with GC and 181 healthy controls, were enrolled in this study. No significant differences were observed in sex, smoking, drinking, current Hp infection, IM, body mass index, serum pepsinogen (PG) I/II ratio, and the degree of neutrophil and monocyte infiltration between subjects with and without FH in GC patients and controls. The proportion of stage I patients was significantly lower in GC patients with FH than in those without FH (69.4% vs. 92.9%, P < 0.001). Furthermore, controls with FH were younger than those without FH (49.45 ± 12.14 vs. 54.27 ± 11.32, P = 0.008) (Table 1).
Aberrant DNA methylation according to Hp infection status and the presence of GC
Methylation levels in six genes were compared according to the presence of GC (GC vs. controls) and Hp infection status (positive vs. negative). For MOS, miR124a-3, NKX6-1, EMX1, CDH1, and TWIST1, methylation levels were higher in patients with GC than in controls. In control subjects, methylation levels for EMX1, CDH1, and TWIST1 were extremely low, demonstrating median PMR values less than 10.
Compared with Hp-negative patients with GC, methylation levels of MOS, miR124a-3, CDH1, and TWIST1 were elevated in Hp-positive GC patients. Among controls, methylation levels of MOS, miR124a-3, EMX1, and CDH1 were significantly elevated in Hp-positive controls compared to levels observed in Hp-negative subjects.
Aberrant DNA methylation according to FH in Hp-positive subjects
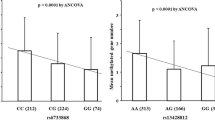
In GC patients with FH, methylation levels of MOS, CDH1, and TWIST1 were lower than those observed in patients without FH (MOS, 29.98 ± 13.62 vs. 50.98 ± 21.23, P < 0.001; CDH1, 13.21 ± 18.00 vs. 33.94 ± 28.17, P < 0.001; TWIST1, 12.77 ± 17.34 vs. 24.00 ± 23.83, P = 0.004) (Fig. 2). In controls with FH, methylation level of MOS were lower than those observed in subjects without FH (32.79 ± 14.24 vs. 39.56 ± 21.76, P = 0.042); however, methylation levels of miR124a-3, NKX6-1, and CDH1 were elevated in controls with FH (miR124a-3, 23.91 ± 23.91 vs. 14.35 ± 13.73, P = 0.006; NKX6-1, 30.15 ± 13.78 vs. 19.25 ± 11.85, P < 0.001; CDH1, 8.51 ± 6.15 vs. 2.45 ± 4.28, P < 0.001) (Fig. 2).
Comparison of MOS (a), miR124a-3 (b), NKX6-1 (c), EMX1 (d), CDH1 (e), and TWIST1 (f) methylation levels between gastric cancer (GC) patients and controls with and without family history (FH) according to H. pylori (Hp) infection. Methylation levels in MOS, CDH1, and TWIST1 are significantly decreased in Hp-positive GC patients with FH when compared with those without FH. MOS methylation levels are significantly decreased in Hp-positive controls; however, methylation levels in miR124a-3, NKX6-1, and CDH1 are significantly increased in Hp-positive controls with FH when compared with those without FH. Bar represents mean of DNA methylation levels. Hp, H. pylori; FH, family history; GC, gastric cancer
Subgroup analysis based on the presence of IM demonstrated similar results. In IM-positive GC patients, methylation levels of MOS, CDH1, and TWIST1 were decreased in subjects with FH compared to those without FH (MOS, 31.03 ± 14.10 vs. 51.64 ± 20.71, P < 0.001; CDH1, 11.68 ± 16.11 vs. 27.63 ± 26.01, P = 0.001; TWIST1, 15.31 ± 18.78 vs. 26.55 ± 25.68, P = 0.025), as well as in IM-negative patient with GC (MOS, 27.08 ± 12.11 vs. 49.47 ± 22.96, p = 0.001; CDH1, 17.46 ± 22.44 vs. 47.95 ± 28.40, P = 0.001; TWIST1, 5.73 ± 9.89 vs. 18.33 ± 18.50, P = 0.018) (Figs. 3 and 4). Among IM-positive controls, miR124a-3, NKX6-1, and CDH1 were differentially methylated in the presence of FH (miR124a-3, 28.84 ± 27.73 vs. 11.40 ± 10.53, P < 0.001; NKX6-1, 30.85 ± 14.18 vs. 17.30 ± 8.20, P < 0.001; CDH1, 9.46 ± 6.14 vs. 3.12 ± 5.05, P < 0.001) (Fig. 3). In the case of IM-negative controls, NKX6-1 and CDH1 were differentially methylated in the presence of FH (NKX6-1, 29.02 ± 13.29 vs. 21.04 ± 14.36, P = 0.039; CDH1, 6.99 ± 5.95 vs. 1.84 ± 3.40, P < 0.001) (Fig. 4).
Comparison of MOS (a), miR124a-3 (b), NKX6-1 (c), EMX1 (d), CDH1 (e), and TWIST1 (f) methylation levels between intestinal metaplasia-positive gastric cancer (GC) patients and controls with and without family history (FH) according to H. pylori (Hp) infection. Among Hp-positive GC patients, methylation levels in MOS, CDH1, and TWIST1 are significantly decreased in patients with FH when compared with those without FH. Among Hp-positive controls, methylation levels in miR124a-3, NKX6-1, and CDH1 are significantly increased in controls with FH when compared with those without FH. Bar represents mean of DNA methylation levels. Hp, H. pylori; FH, family history; GC, gastric cancer
Comparison of MOS (a), miR124a-3 (b), NKX6-1 (c), EMX1 (d), CDH1 (e), and TWIST1 (f) methylation levels between intestinal metaplasia-negative gastric cancer (GC) patients and controls with and without family history (FH) according to H. pylori infection. Among Hp-positive GC patients, methylation levels in MOS, CDH1, and TWIST1 are significantly decreased in patients with FH when compared with those without FH. Among Hp-positive controls, methylation levels in NKX6-1 and CDH1 are significantly increased in controls with FH when compared with those without FH. Bar represents mean of DNA methylation levels. Hp, H. pylori; FH, family history; GC, gastric cancer
In subgroup analysis by histology, methylation levels of MOS, CDH1, and TWIST1 were decreased in subjects with FH than those without FH among intestinal-type GC (MOS, 29.97 ± 13.50 vs. 48.32 ± 20.31, P < 0.001; CDH1, 11.83 ± 17.37 vs. 33.77 ± 29.09, P < 0.001; TWIST1, 12.46 ± 16.97 vs. 24.81 ± 25.62, P = 0.018) (Fig. 5). Methylation levels of MOS and CDH1 were lower in subjects with FH than in those without FH among diffuse-type GC (MOS, 30.43 ± 15.34 vs. 55.82 ± 19.33 P < 0.001; CDH1, 13.74 ± 18.47 vs. 36.17 ± 27.32, P = 0.006) (Fig. 5).
Comparison of MOS (a), miR124a-3 (b), NKX6-1 (c), EMX1 (d), CDH1 (e), and TWIST1 (f) methylation levels between gastric cancer (GC) patients and controls with and without family history (FH) according to H. pylori infection and histology (intestinal vs diffuse). Among intestinal type GC patients with Hp infection, methylation levels in MOS, CDH1, and TWIST1 are significantly decreased in patients with FH when compared with those without FH. Among diffuse type GC patients with Hp infection, MOS and CDH1 methylation levels are significantly decreased in patients with FH compared with those without FH. Bar represents mean of DNA methylation levels. Hp, H. pylori; FH, family history; GC, gastric cancer
Aberrant DNA methylation according to FH in Hp-negative subjects
In Hp-negative GC and controls, MOS, miR124a-3, NKX6-1, EMX1, CDH1, and TWIST1 were not differentially methylated between subjects with and without FH (Fig. 2). In subgroup analysis by IM and histology, no significant differences were found.
Changes in DNA methylation levels over time
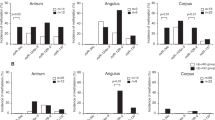
Figure 6 reveals changes in the levels of DNA methylation over time in Hp-eradicated subjects. It presented as the mean values of collected data at each interval. The average follow-up duration was 57.2 months (range 2–150 months). In patients with GC, a significant decrease in CDH1 methylation was observed 2 years after Hp eradication (2 years, P < 0.001; 3–4 years, P < 0.001; ≥ 5 years, P = 0.014); in controls, CDH1 methylation was significantly decreased 3–4 years after eradication (3–4 years, P = 0.025; ≥ 5 years, P = 0.01) (Fig. 6). The levels of MOS, miR124a-3, NKX6-1, EMX1, and TWIST1 did not significantly change after Hp eradication in patients with GC, as well as controls (Fig. 6).
Changes in DNA methylation levels over time in the MOS (a), miR124a-3 (b), NKX6-1 (c), EMX1 (d), CDH1 (e), and TWIST1 (f) genes in Helicobacter pylori (Hp)-eradicated gastric cancer (GC) patients and controls according to the presence of family history (FH). The CDH1 methylation level is significantly decreased in GC patients at 2 years after Hp eradication (2 years, P < 0.001; 3–4 years, P < 0.001; ≥ 5 years, P = 0.014) and in controls at 3–4 years after eradication (3–4 years, P = 0.025; ≥ 5 years, P = 0.01) when compared with baseline levels. Methylation levels of CDH1 were significantly decreased from 2 years after Hp eradication (2 years, P = 0.001; 3–4 years, P = 0.001; 5 ≥ years, P = 0.032) after Hp eradication than baseline levels in GC patients without FH, and 2 (P = 0.030) and 3–4 years (P < 0.001) in GC patients with FH, 2 years (P = 0.022) in controls without FH, and 3–4 (P = 0.001), and 5 ≥ years (P = 0.001) in controls without FH. Significant decreases in methylation levels of MOS, miR124a-3, and TWIST1 were observed at 2 years (miR124a-3, P = 0.008; TWIST1, P = 0.008) and 5 ≥ years (MOS, P = 0.008; miR124a-3, P = 0.005) after eradication in GC patients with FH. DNA methylation results are presented as mean ± standard error. Hp, H. pylori; FH, family history; GC, gastric cancer
Following subgroup analysis according to the presence of FH, CDH1 methylation levels demonstrated similar trends. CDH1 methylation levels were significantly decreased at 2 (P < 0.001), 3–4 (P = 0.002), and ≥ 5 years (P = 0.001) after Hp eradication observed in GC patients without FH, and 2 (P = 0.04) and 3–4 years (P = 0.002) after eradication in GC patients with FH, and 2 years (P = 0.035) after eradication in controls without FH, and 3–4 (P < 0.001) and ≥ 5 years (P = 0.001) after eradication in controls with FH when compared with baseline levels (Fig. 6). Additionally, methylation levels of miR124a-3 were significantly decreased at 2 (P = 0.002) and ≥ 5 years (P < 0.001) after eradication in GC patients with FH when compared with baseline levels (Fig. 6).
When subjects with FH and without FH were compared, significant differences in the methylation levels of MOS were constantly observed at 1 (P = 0.023), 3–4 (P = 0.005), and ≥ 5 years (P = 0.001) after eradication in GC patients, and 2 (P = 0.035), 3–4 (P = 0.014), and ≥ 5 years (P = 0.048) after eradication in controls (Fig. 6). However, significant differences in the methylation levels of CDH1 and TWIST1 in GC patients disappeared from 1 year after Hp eradication (Fig. 6). The difference in miR124a-3, NKX6-1 and CDH1 methylation levels between controls with and without FH were not observed from 1 year after Hp eradication (Fig. 6).
Discussion
In the present study, the methylation status was compared between subjects with and without FH to identify specific genes associated with GC susceptibility in the presence of FH. We observed that methylation levels of MOS, CDH1, and TWIST1 decreased in Hp-positive GC patients with FH when compared with those without FH. In Hp-positive healthy controls, MOS, miR124a-3, NKX6-1, and CDH1 were differentially methylated between subjects with and without FH. Additionally, CDH1 methylation levels decreased to certain levels following Hp eradication in both GC patients with and without FH.
Regarding the six genes examined in this study, it has been previously demonstrated that hypermethylation of these genes in noncancerous gastric mucosa is associated with GC occurrence [3, 25,26,27,28]. Additionally, these genes are linked to Hp-induced DNA methylation in GC development [3, 25]. However, to date, evidence demonstrating the relationship between aberrant DNA methylation and FH of GC remains scarce. Kim et al. have reported that methylation of helicase-like transcription factor (HLTF), determined by methylation-specific PCR, is significantly associated with early GC patients with FH, suggesting that HLTF methylation could serve as a susceptible marker to assess the risk of GC in individuals with FH [29]. Chang et al. have revealed that ROBO2 gene methylation, as measured by methylation-specific PCR, is significantly lower in patients with FH of GC than in patients without FH [30]. We observed a decrease in methylation levels of MOS, CDH1, and TWIST1 in GC patients with FH when compared with those without FH. Our results suggest that MOS, CDH1, and TWIST1 gene methylations may be associated with sporadic gastric tumorigenesis, and epigenetic silencing of MOS, CDH1, and TWIST1 may emerge after the development of GC rather than before GC development, or during the formation of an epigenetic field.
In the present study, subjects with and without FH demonstrated similar inflammation-associated characteristics, such as the presence of IM, the degree of neutrophil and monocyte infiltration, and serum PG I/II ratio. Despite previous findings suggesting that IM is associated with aberrant DNA methylation levels [31], our findings demonstrating decreased methylation levels of MOS, CDH1, and TWIST1 in GC patients with FH persisted after adjustment for IM. Additionally, analyses of NKX6-1 and CDH1 revealed significant differences between controls with and without FH after adjustment for IM. However, we did not detect any association of FH with genetic DNA methylation levels among those without current Hp infections. The small number of subjects with true negative Hp infection could be insufficient to prove significance. The negative Hp infection could have hindered the detection of an association with aberrant DNA methylation.
We demonstrated that, compared with baseline levels, methylation levels of CDH1 decreased constantly 2 years after Hp eradication in patients with GC and 3–4 years after eradication in controls. Among patients with GC, CDH1 methylation levels did not completely disappear for more than 5 years, and remained persistently higher than levels of control. Similar trends in changes of CDH1 methylation levels were found in subjects with and without FH. In GC patients with FH, methylation levels of MOS, miR124a-3, and TWIST1 decreased 2 and/or ≥ 5 years after eradication, suggesting that the changes in methylation levels could differ according to the presence of FH. Although our study did not demonstrate that methylation levels of miR124a-3, NKX6-1, EMX1, and TWIST1 decreased constantly beginning 1 year after Hp eradication, it should be stated that these levels did not significantly increase when compared with baseline levels at any interval. Notably, methylation levels of MOS in the Hp-eradicated GC patients and controls remained persistently higher than baseline levels. Additionally, the significant difference in the miR124a-3, NKX6-1, CDH1 and TWIST1 methylation levels between subjects with and without FH disappeared after Hp eradication though MOS methylation levels in subjects with FH were constantly decreased than those in subjects without FH after eradication. Based on our findings, as well as previous studies, Hp eradication could prevent GC, which could be attributed to the amelioration of chronic inflammation and atrophy, as well as to the prevention of further progression of an epigenetic field for cancerization and restoration of epigenetic alterations. Residual methylation in the gastric mucosa after eradication might be relevant for GC development after successful Hp eradication. Furthermore, these findings might support one of the molecular mechanisms explaining the results of previous studies reporting that Hp eradication reduces the incidence of metachronous GC [32].
Although several studies have demonstrated changes in DNA methylation after Hp eradication, most studies on aberrant methylation were followed up within 1 year of treatment [9, 10]. A previous report has shown that CDH1 methylations levels decreased at 6 weeks [10] and 1 year after Hp eradication [9]. Recently, a 5-year follow-up study has indicated that the CDH1 methylation rate decreased gradually over time after Hp eradication in both non-IM and IM, but only 78 patients including 27 GC patients were enrolled [33]. Our study included total 382 subjects and followed 60 GC patients and 72 controls over 5 years. Furthermore, several previous studies have utilized methylation-specific PCR, but it does possess some limitations, including several false-positive results and provides a qualitative analysis [34]. In this study, the strengths included the adoption of quantitative methods, MethyLight in the evaluation of DNA methylation, and enrollment of both Hp-negative and Hp-positive subjects, as well as analysis of subjects considering the presence of IM and histology.
In summary, our results suggest that MOS and CDH1 methylation can provide a link between aberrant DNA methylation and GC with FH, and might be a useful marker for the risk of GC development. Hp eradication was associated with a significant decrease in CDH1 in GC patients and controls, and significant difference of CDH1 methylation levels among subjects with and without FH disappeared in Hp-eradicated GC patients and controls. Further prospective and long-term follow-up studies with larger patient groups are required to confirm the difference of the these gene methylation levels among subjects with and without FH, and the direct effect of Hp eradication on the methylation changes in various genes in gastric epithelial cells.
References
Ushijima T. Epigenetic field for cancerization. J Biochem Mol Biol. 2007;40:142–50.
Ushijima T, Sasako M. Focus on gastric cancer. Cancer Cell. 2004;5:121–5.
Chan AO, Lam SK, Wong BC, Wong WM, Yuen MF, Yeung YH, et al. Promoter methylation of E-cadherin gene in gastric mucosa associated with Helicobacter pylori infection and in gastric cancer. Gut. 2003;52:502–6.
Niwa T, Tsukamoto T, Toyoda T, Mori A, Tanaka H, Maekita T, et al. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res. 2010;70:1430–40.
Maekita T, Nakazawa K, Mihara M, Nakajima T, Yanaoka K, Iguchi M, et al. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006;12:989–95.
Shin CM, Kim N, Jung Y, Park JH, Kang GH, Park WY, et al. Genome-wide DNA methylation profiles in noncancerous gastric mucosae with regard to Helicobacter pylori infection and the presence of gastric cancer. Helicobacter. 2011;16:179–88.
Shin CM, Kim N, Jung Y, Park JH, Kang GH, Kim JS, et al. Role of Helicobacter pylori infection in aberrant DNA methylation along multistep gastric carcinogenesis. Cancer Sci. 2010;101:1337–466.
Leung WK, Man EP, Yu J, Go MY, To KF, Yamaoka Y, et al. Effects of Helicobacter pylori eradication on methylation status of E-cadherin gene in noncancerous stomach. Clin Cancer Res. 2006;12:3216–21.
Perri F, Cotugno R, Piepoli A, Merla A, Quitadamo M, Gentile A, et al. Aberrant DNA methylation in non-neoplastic gastric mucosa of H. pylori infected patients and effect of eradication. Am J Gastroenterol. 2007;102:1361–71.
Nakajima T, Enomoto S, Yamashita S, Ando T, Nakanishi Y, Nakazawa K, et al. Persistence of a component of DNA methylation in gastric mucosae after Helicobacter pylori eradication. J Gastroenterol. 2010;45:37–44.
Yaghoobi M, Bijarchi R, Narod SA. Family history and the risk of gastric cancer. Br J Cancer. 2010;102:237–42.
Oliveira C, Pinheiro H, Figueiredo J, Seruca R, Carneiro F. Familial gastric cancer: genetic susceptibility, pathology, and implications for management. Lancet Oncol. 2015;16:e60–70.
Shin CM, Kim N, Yang HJ, Cho SI, Lee HS, Kim JS, et al. Stomach cancer risk in gastric cancer relatives: interaction between Helicobacter pylori infection and family history of gastric cancer for the risk of stomach cancer. J Clin Gastroenterol. 2010;44:e34–e3939.
Choi YJ, Kim N, Jang W, Seo B, Oh S, Shin CM, et al. Familial clustering of gastric cancer: a retrospective study based on the number of first-degree relatives. Medicine. 2016;95:e3606.
El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, Young HA, et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398–402.
Kim HJ, Kwon M, Kim N, Lee JB, Won S. The influence of family history on stage and survival of gastric cancer according to the TGFB1 C-509T polymorphism in Korea. Gut Liver. 2020;14:79–88.
Yoon H, Kim N, Shin CM, Lee HS, Kim BK, Kang GH, et al. Risk factors for metachronous gastric neoplasms in patients who underwent endoscopic resection of a gastric neoplasm. Gut liver. 2016;10:228–36.
Shin CM, Kim N, Lee HS, Park JH, Ahn S, Kang GH, et al. Changes in aberrant DNA methylation after Helicobacter pylori eradication: a long-term follow-up study. Int J Cancer. 2013;133:2034–42.
Maeda M, Nakajima T, Oda I, Shimazu T, Yamamichi N, Maekita T, et al. High impact of methylation accumulation on metachronous gastric cancer: 5-year follow-up of a multicentre prospective cohort study. Gut. 2017;66:1721–3.
Kaise M, Yamasaki T, Yonezawa J, Miwa J, Ohta Y, Tajiri H. CpG island hypermethylation of tumor-suppressor genes in H. pylori-infected non-neoplastic gastric mucosa is linked with gastric cancer risk. Helicobacteria. 2008;13:35–41.
Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39.
Puisieux A, Valsesia-Wittmann S, Ansieau S. A twist for survival and cancer progression. Br J Cancer. 2006;94:13–7.
Kokkola A, Kosunen TU, Puolakkainen P, Sipponen P, Harkonen M, Laxen F, et al. Spontaneous disappearance of Helicobacter pylori antibodies in patients with advanced atrophic corpus gastritis. APMIS. 2003;111:619–24.
Trinh BN, Long TI, Laird PW. DNA methylation analysis by MethyLight technology. Methods. 2001;25:456–62.
Ando T, Yoshida T, Enomoto S, Asada K, Tatematsu M, Ichinose M, et al. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int J Cancer. 2009;124:2367–74.
Nanjo S, Asada K, Yamashita S, Nakajima T, Nakazawa K, Maekita T, et al. Identification of gastric cancer risk markers that are informative in individuals with past H. pylori infection. Gastric Cancer. 2012;15:382–8.
Kang GH, Lee S, Cho NY, Gandamihardja T, Long TI, Weisenberger DJ, et al. DNA methylation profiles of gastric carcinoma characterized by quantitative DNA methylation analysis. Lab Invest. 2008;88:161–70.
Shin CM, Kim N, Park JH, Kang GH, Kim JS, Jung HC, et al. Prediction of the risk for gastric cancer using candidate methylation markers in the non-neoplastic gastric mucosae. J Pathol. 2012;226:654–65.
Kim JJ, Chung SW, Kim JH, Kim JW, Oh JS, Kim S, et al. Promoter methylation of helicase-like transcription factor is associated with the early stages of gastric cancer with family history. Ann Oncol. 2006;17:657–62.
Chang ZW, Dong L, Qin YR, Song M, Guo HY, Zhu QL. Correlations between gastric cancer family history and ROBO2 and RASSF2A gene methylations. J Cancer Res Ther. 2016;12:597–600.
Kang GH, Shim YH, Jung HY, Kim WH, Ro JY, Rhyu MG. CpG island methylation in premalignant stages of gastric carcinoma. Cancer Res. 2001;61:2847–51.
Choi IJ, Kook MC, Kim YI, Cho SJ, Lee JY, Kim CG, et al. Helicobacter pylori therapy for the prevention of metachronous gastric cancer. N Engl J Med. 2018;378:1085–95.
Michigami Y, Watari J, Ito C, Nakai K, Yamasaki T, Kondo T, et al. Long-term effects of H. pylori eradication on epigenetic alterations related to gastric carcinogenesis. Sci Rep. 2018;8:14369.
Kristensen LS, Hansen LL. PCR-based methods for detecting single-locus DNA methylation biomarkers in cancer diagnostics, prognostics, and response to treatment. Clin Chem. 2009;55:1471–83.
Funding
This work was supported by a Grant (2011-0030001) from the National Research Foundation (NRF) for the Global Core Research Center (GCRC) funded by the Ministry of Science, ICT and Future Planning (MSIP), Republic of Korea.
Author information
Authors and Affiliations
Contributions
HJK analyzed the data and drafted the article; NK designed the protocol, enrolled participants, collected data, and edited the manuscript; HWK analyzed data; JHP performed the molecular biological study; CMS and DHL interpreted the data and edited the manuscript. All authors have read and approved the final draft of this paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no confict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kim, H.J., Kim, N., Kim, H.W. et al. Promising aberrant DNA methylation marker to predict gastric cancer development in individuals with family history and long-term effects of H. pylori eradication on DNA methylation. Gastric Cancer 24, 302–313 (2021). https://doi.org/10.1007/s10120-020-01117-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-020-01117-w