Abstract
With the rapid development of information technologies, multi-source heterogeneous data has become an open problem, and the data is usually modeled as graphs since the graph structure is able to encode complex relationships among entities. However, in practical applications, such as network security analysis and public opinion analysis over social networks, the structure and the content of graph data are constantly evolving. Therefore, the ability to continuously monitor and detect interesting patterns on massive and dynamic graphs in real-time is crucial for many applications. Recently, a large group of excellent research works has also emerged. Nevertheless, these studies focus on different updates of graphs and apply different subgraph matching algorithms; thus, it is desirable to review these works comprehensively and give a thorough overview. In this paper, we systematically investigate the existing continuous subgraph matching techniques from the aspects of key techniques, representative algorithms, and performance evaluation. Furthermore, the typical applications and challenges of continuous subgraph matching over dynamic graphs, as well as the future development trends, are summarized and prospected.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Over the last decade, information networks such as social networks, communication networks, world wide web, financial transaction networks, etc., have become ubiquitous and pervasive. Apart from their increasing data scale and data types, one of the most important aspects is that the structure and content of the graphs constantly evolve due to the frequent updates in the real world. Taking social networks as an example, there are over 1.4 billion daily active users on Facebook, generating more than 500,000 posts/comments and 4 million likes per minute, leading to a flood of updates on the Facebook social network [2]. In addition, Google+ added 10 million new users in the two weeks since its launch in 2011 [32]. Therefore, the ability to continuously monitor and detect patterns of interest is essential for obtaining meaningful and up-to-date discoveries in such frequently updated graphs. Meanwhile, this monitoring capability is required for many applications from a variety of domains.
-
Social networks. The applications may involve: 1) targeted advertising, spam or fraudulent activities detection [14, 38, 74] and 2) fake news propagation monitoring [66].
-
Protein-interaction network. In a protein-interaction network, by matching a known protein network with a dynamic protein interaction network, the mutated protein structure can be found quickly from the protein interaction network [19, 77].
-
Transportation networks. In transportation networks, we can monitor the real-time traffic accidents by matching the pattern graph with the dynamic road network data graph [66].
-
Computer networks. In dynamic computer networks, based on the continuous subgraph matching technique, we can monitor cyber-attack events or detect anomaly flow in time [17].
-
Knowledge graphs. It can deal with evolving knowledge over time to support continuous question answering [13] and reasoning over RDF graph is also based on such pattern monitoring [6].
Given a dynamic data graph and a query graph, the continuous subgraph matching problem can be described as identifying and monitoring the query graph in the dynamic data graph continuously and reporting the newer up-dates of the matching results. Matching here refers to the same structure and meeting the specific semantic constraint. Now, we take cyber-attack event detection and credit card fraud detection as examples to illustrate how to abstract an event into a pattern graph and the significance of continuous subgraph matching.
Example 1
Figure 1a demonstrates a graph-based description of cyber-attack pattern. A victim browses the compromised website, causing a downloading of malware scripts. Then, the malware scripts establish a communication between the victim and the botnet command and control server. When the victim registers at the botnet command and control server, he receives a command at the same time, leading to an information exfiltration back to the botnet command and control server. Obviously, if we can detect the attack pattern (based on the subgraph matching) in the network, potential malicious activities could be avoided. Recently, Verizon, an American communications company, analyzed 100,000 security incidents over the past decade and found that 90% of them could be easily classified into ten major graph-based attack patterns [7].
Example 2
Figure 1b shows an example of a credit card fraud with a series of transactions modeled by a pattern graph. Criminals try to cash out illegally by making bogus deals with merchants and middlemen. They first establish a credit payment to the merchant; When the merchant receives the real payment from the bank, he transfers the money to the middleman account(s), which further transfers the money back to the criminal to complete the cashing. Apparently, this pattern also can be easily modeled as a query graph. By detecting this pattern query in the financial transaction networks, fraud can be avoided effectively.
Challenges and Limitations. Owing to the importance and practicality of continuous subgraph matching, how to effectively and efficiently find the matching results of a given query graph continuously over the dynamic graphs is a meaningful research topic in the era of big data. By observing its practical application requirements, we identify a list of challenges that the continuous subgraph matching may face:
-
Frequent data updates. The graph data in the network system, such as social networks, data center networks, or financial networks, updates all the time. Thus, traditional static graph matching techniques would need to rebuild an index and rematch the whole graph data for each update, which is a time-consuming and labor-intensive approach.
-
Large-scale data. Due to the constant updates, the amount of dynamic graph data is larger than that of static graph data, increasing the difficulty of graph subgraph matching.
-
Increased demand for real-time analysis. In many real-time analysis applications, whenever the graph is updated, the new matching result requires to be obtained in time; otherwise, the application value of the matching result will be reduced or lost. For example, in network security, real-time monitoring of suspicious data transmission modes is required, and a delay in analyzing the matching results can result in network paralysis.
The primary motivation for conducting this survey is twofold. Firstly, a lot of graph algorithms have been proposed to perform continuous subgraph matchings tasks in recent years. However, existing surveys are somewhat out-of-date, and no new research and recommendations are discussed. The latest survey [39] was published five years ago, and there was only a brief introduction to subgraph matching in the dynamic graph. Secondly, the surveys [33] and [46] only introduce and summarize the existing algorithm from a certain aspect, not comprehensive. Additionally, existing survey [47] solely focus on the subgraph matching algorithms in static graph [36, 63, 85, 87]. These existing surveys did not provide comprehensive reviews on continuous subgraph matching over the dynamic graph data. In particular, Our survey is the first one to illustrate the existing research effort and progress that has been made in the area of continuous subgraph matching in the dynamic graph regarding key techniques, representative algorithms, and performance evaluations. Meanwhile, there are some related surveys, such as pattern mining [28] and community search [27]. However, the former focuses on graph mining in dynamic graphs while the latter focuses on community retrieval over big graphs. Although both of these technologies require subgraph matching algorithms, it is not the focus of their technical optimization.
In this survey, we introduce the existing continuous subgraph matching methods which committed to solving the above problems. First, we categorize the research of continuous subgraph matching systematically according to the different practical application requirements. And then, we discuss two different types of continuous subgraph matching approaches based on the update methods of the dynamic graph. Among them, continuous subgraph matching for graph structure change is the most commonly study, and the incremental matching approach is the most advanced method applied to solve this problem, which attempts to identify changes to previously matched data in response to updated dynamic graph data to meet the increased demand for the real-time analysis.
The rest of the paper is organized as follows. Section 2 provides a detailed definition of continuous subgraph matching and classification. Sections 3 and 4 describe the concepts of and matching techniques for structure-based and content-based changes, respectively. Section 5 compares the performance of different matching algorithms. Section 6 introduces the current applications of continuous subgraph matching. Section 7 presents a list of future topics, and Sect. 8 presents the conclusion.
2 Definition and classification of continuous subgraph matching
In this section, we first formally introduce continuous subgraph matching and then have a comprehensive classification of continuous subgraph matching from a variety of different methodological perspectives.
2.1 Preliminary definition
The graph is usually is defined as (V, E, L), where V is a set of vertices, E is a set of edges between, and L is a label function that associates each vertex v \(\in \) V with a set of labels. Any edge \(e \in E_G\) in the graph can be noted as \((v_i, v_j)\), where \(v_i, v_j \in V_G\). In this paper, we use \(G=(V_G,E_G,L_G)\) to represent data graph, and \(P=(V_P,E_P,L_P)\) to represent pattern graph. Currently, there are two data graph types: super-large graphs, such as social networks, or a combination of many small graphs, such as the AIDS Antiviral dataset [5] that comprises numerous small pictures showing the atomic structure of chemicals. As the current continuous subgraph matching problem mainly relates to super-large graphs, this paper concentrates on continuous subgraph matching for super-large graphs.
A dynamic graph, also known as a streaming graph, is a graph that evolves over time. There are two categories of dynamic graph updates, namely (1) graph structure updates, where the vertices and edges in the graph are inserted or deleted over time, causing changes in the graph structure; (2) graph content updates, where the content or attributes of the data objects in the graph are updated, resulting in content change.
The definition of the dynamic graph is given in the next section. As the current research is mainly focused on the structural update of the data graph, the definition of the dynamic graph is given in the graph structure update.
2.1.1 Graph structure update
Definition 1
(Graph Update Stream) A graph update stream \(\Delta g\) is construct with a sequence of update operations \((\Delta g_1,\Delta g_2,\cdots )\), where \(\Delta g_1\) is a triple \(\langle op, v_i,v_j\rangle \) such that \(op=\{I,D\}\) is the type of operations, with I and D representing edge insertion and edge deletion respectively of an edge \((v_i,v_j)\).
A dynamic graph abstracts an initial graph \(G\) \(=\) \((V_G,E_G,L_G)\) and an update stream \(\Delta g\). After applying \(\Delta g\) to G, it transforms to \(G'\). Note that insertion operation creates new edges between vertices and could also add new vertices. The same does for deletion operations.
Definition 2
(Subgraph isomorphism) Given two graphs \(G_1\)=\((V_1,E_1,L_1)\) and \(G_2\)=\((V_2,E_2,L_2)\), an embedding of \(G_1\) in \(G_2\) (or, from \(G_1\) to \(G_2\)) is an bijective function f: (1) \(\forall v \in V_1\), \(L_1(v)=L_2(f(v))\); and (2) \(\forall (v_1, v_2)\in E_1\), \((f(v_1),f(v_2))\in E_2\).
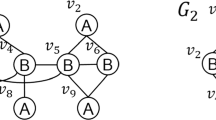
Example 3
In Fig. 2, the dashed box shows the subgraph \(M=\) \([(v_1,u_3)\),\((v_2,\) \(u_6)\),\((v_3,u_5)\),\((v_4,u_8)]\) that matches the pattern graph P after the initial data graph G is changed to \(G_1\) with the update operation \(\Delta g=\{\langle I,u_5,u_8\rangle \}\).
Recently, graph functional dependencies [25], keys [21], and association rules [24] are all defined based on subgraph isomorphic matching [62]. However, it is a NP-complete problem due to the exponential search space resulting generated by all possible subgraphs [65] [35]. Subgraph isomorphism often imposes stronger topological constraints on the matching process to obtain more consistent matching results. Some applications, such as social networks, do not require high matching accuracy, which greatly restricts matching efficiency. In view of its intractability, approximate matching has been studied to obtain inexact solutions, which allows vertex/edge mismatching [9, 70]. Differs from approximate matching, simulation matching does not allow vertex/edge mismatching. The simulation matching is described in more detail below.
According to the degree of the matching approximations, it is divided into Bounded simulation [23], Graph simulation [53], Dual simulation [50], Strong simulation [50] and Strict simulation, all above are collectively called simulation matching. Bounded simulation matching relaxes the strict structural constraints of isomorphism matching by moving from edge-to-edge mapping to edge-to-path mapping. If a data vertex u matches a query vertex v, they only need to satisfy that \(L_P(u)\)=\(L_G(v)\) and \(L_P(u')\)=\(L_G(v')\), where \(u'\) is one of the descendant vertex of u and \(v'\) is one of the child vertex have of \(v'\). Compared with bounded simulation, graph simulation requires that \(u'\) is one of the child vertexes of u. It means that vertex u maintains the same successor relationship with the corresponding vertex v. The dual simulation further requires u to maintain the same precursor relationship with v. On the basis of dual simulation, strong simulation requires that the radius of the subgraph containing the matching vertex(the subgraph may contain non-matching vertices) is not larger than the diameter of the pattern graph. Strict simulation matching is the strictest one of simulation matching, and it further requires the subgraph composed entirely of the matching vertices.
Note that simulation matching can tolerate some noise and errors in matching results. Thus, it plays an important role in some applications such as social network and Web network analysis. Although differences between the results of simulation matching and subgraph isomorphism remain, simulation matching results usually meet the needs of practical applications. Hence, it is still regarded as the correct matching result. Take graph simulation as an example, and it is defined as follows:
Definition 3
(Graph simulation) Given a pattern graph \(P=(V_P,E_P,L_P)\) and \(G=(V_G,E_G,L_G)\), graph simulation refers that there is a binary relationship \(S\) \(\subseteq \) \(V_P\) \(\times \) \(V_G\) between P and G, which satisfies that:
-
(1)
\(\forall \) \((v, u)\) \(\subseteq \) \(S\), \(L_G(v)\)=\(L_P(u)\).
-
(2)
\(\forall \) \(v\) \(\in \) \(V_P\), we have: (a) \(\exists \) \(u\) \(\in \) \(V_G\) with \((v,u)\) \(\in \) \(S\), and (b) \(\forall \) \((v, v)\) \(\in \) \(E_G\), \(\exists \) \((u,u')\) \(\in \) \(E_G\) with \((v',u')\) \(\in \) \(S\).
Example 4
As shown in Fig. 3, the vertex \(u_2\) in the initial data graph G has the same label as \(v_2\) in the pattern graph P. Moreover, \({u_2}'s\) successor \(u_4\) has the same label as \({v_2}'s\) successor \(v_4\). Therefore, we can infer that \(u_2\) matches \(v_2\). Similarly, because \({v_3}'s\) successor in the pattern graph P is empty, we can infer that \(v_3\) matches \(u_3\). As there is no vertex in \({u_4}'s\) successor and its label is the same as \({v_4}'s\) successor, \(v_3\) , \(u_4\) and \(v_4\) do not match. Therefore, according to the definition of graph simulation matching, for \(t=1\), after the initial data graph G was updated with the operation \(GC_2=\{\langle I, u_4, u_6\rangle \}\), \(u_4\) and \(v_4\) met the matching conditions. Furthermore, the subgraphs \(M_1=[(v_2, u_2), (v_3, u_3), (v_4, u_4)]\) and \(M_2=[(v_2, u_2), (v_3, u_6), (v_4, u_4)]\) matched the pattern graph generated. Note that, although the structure of \(M_2\) is different from that of the pattern graph P, it is still considered as a match.
2.1.2 Graph content update
Graph content update means that the labels of the vertex/edge or specific object evaluation in the graph can change with time.
Example 5
As shown in Fig. 4, for time \(t=1\), the label of \(u_3\) and \(u_6\) in the initial data graph G were changed. The dotted box shows the subgraph \(M = [(v_1, u_3), (v_2, u_6), (v_3, u_5), (v_4, u_8)]\), which matches the pattern graph after the data graph is updated.
2.2 Classification of continuous subgraph matching
In this subsection, we classify continuous subgraph matching according to five methodological perspectives and introduce the representative algorithms for each class. Since each classification is not independent, there are intersections between the algorithms for each classification.
2.2.1 Dynamic feature of data graph
From the dynamic feature of data graph, continuous subgraph matching is divided into structural change and content change based on whether the topological structure of the graph is changed. Currently, many studies about continuous subgraph matching mainly focus on structural changes caused by the dynamic addition and deletion of vertices or edges in the data graph. It is mainly used in network anomaly attack detection and social network relationship detection. Continuous subgraph matching for structure change was first proposed by Wang et al. [75] in 2009. They constructed a node-neighbor tree (NNT) to filter the matching candidate set, which can effectively reduce the negative matching results. Later, some representative algorithms are proposed, including IncIsoMatch [23], SJ-Tree [16], graph simulation (DDST [66], and IncBMatch [23]), which further improve the execution efficiency of subgraph matching.
Content changes for graph mainly appears in the data center. Many studies have abstracted the network topological structure of the data center into a data graph for data analysis, where each vertex in the graph represents a server, and the edges between vertices represent the links between servers. In this case, the topological structure of the graph data does not change frequently, but the labels of the vertices (i.e., the amount of free memory of the server) and edges (i.e., the effective bandwidth of the link) in the data graph change frequently over time. In order to deal with this situation effectively, BoZhong et al. [88] proposed the Gradin algorithm. It put forward a N-dimensional grid index for tracking the evolution data and can also be directly applied to the frequently updated edge labels in a data graph.
2.2.2 Strength of consistency constraint
From the strength of consistency constraint, according to whether a bijective function or binary relationship is used, continuous subgraph matching can be divided into subgraph isomorphism and simulation matching, respectively. Subgraph isomorphism guarantees that the matching results are completely consistent with the pattern graph. It is mainly used for protein-interaction network analyzing [39], network abnormal behavior monitoring [17] and other data analysis applications with strict structural requirements.
While simulation matching obtains the results via binary relationship, and the matching results are approximate matches. First, it generates a matching candidate set of each query vertex according to the label of the data vertex and then filters out unmatched vertices according to the different approximate matching degrees of the precursor and successor of the vertex in the pattern graph. Representative algorithms include DDST [66] and IncBMatch [23], etc. Particularly, the results obtained by the IncBMatch algorithm satisfy the matching conditions even if the structure of the subgraphs is different from the pattern graph. Meanwhile, the matching process of IncBMatchcan is performed in polynomial time with higher efficiency. Simulation matching is more flexible, improves the matching efficiency, and identifies more useful matching results. Hence, it is mainly used for monitoring traffic accidents on the road network [66], and detecting the relationship between people and various groups [23] (such as the drug transaction relationship network). This graph data analysis application focuses more on mining the relationship between vertices.
2.2.3 Type of matched results
From the type of matched results, a continuous subgraph matching algorithm is classified as an exact algorithm or approximate algorithm due to the accuracy of the obtained matching results. Exact algorithms ensure that the matching results are completely accurate and are mainly used in many fields such as network anomaly detection and biological data analysis, which rely on accurate matching results. The examples of exact algorithms used to match data graphs and pattern graphs include IncIsoMatch [23], SJ-Tree [16], and Gradin [88].
However, many applications that use real-time results require that all matching results be returned quickly, making complicated exact matching algorithms unsuitable. Coincidently, these applications could also tolerate some vertex/edge mismatches (i.e., false-positive results). To address these problems, the approximate subgraph matching algorithm is generated. It is different from simulation matching as it is usually based on mathematical models such as probability statistics. The error ratio of the matching results can be kept within a certain range through parameter adjustment. All research works which presented NNT [75], Replication mechanism [55], and SSD [30], proposed an approximate graph matching algorithm that converts the graph matching problem into a more easily solved problem, trading matching accuracy for matching efficiency.
Note that the exact algorithm and approximate algorithm are two algorithms that determine the accuracy of matching results, while subgraph isomorphism and simulation matching are different types of graph matching models.
2.2.4 Computational environment
From the computational environment, continuous subgraph matching can either be centralized or distributed according to whether it is deployed on a distributed platform or single machine. Centralized continuous subgraph matching runs on a single computer with either join-based or exploration-based matching methods and mainly handles small-scale graph data. The join-based matching method decomposes the query graph, matches each query fragment, and then connects the obtained matching results. Generally, this method needs to build an index. The exploration-based method [69] starts from a vertex in the data graph and explores the entire data graph according to the query graph’s structure. Differ from join-based matching. This method does not need to build an index.
Distributed continuous subgraph matching mainly depends on a distributed parallel graph processing framework to process dynamic graph data with a large scale and high computational complexity. Currently, there are three types of mainstream distributed parallel graph computing systems, namely vertex-centric model (including Pregel [51], Trinity [64], Giraph [1], and GraphLab [49]), block-centric model (Blogel [78], BLADYG [11]), and auto-parallelization model (GRAPE [26]). Among them, vertex-centric and block-centric models need to modify the algorithm according to the characteristics of the model, which is difficult for the unfamiliar user to recast the parallel models. In contrast, the auto-parallelization model does not need to require that; it simply requires the user to provide three sequential (incremental) algorithms for the data graph with a small amount of content. The logic of the existing algorithm does not need to be modified. Additionally, all the three distributed parallel graph computing systems follow the BSP model [72] and the exploration-based approach is generally adopted for distributed matching owing to the need for the incremental updating of matching results.
Table 1 shows the performance of different types of distributed parallel graph computing systems in terms of shortest path query. Using the US road network as a data set, it can be found that the performance of the GRAPE system is significantly better than other types of systems.
2.2.5 Dependency relationship of queries
From the dependency relationship of queries, continuous subgraph matching can also be divided into single query answer or multi-query answer according to whether it expresses the continuous subgraph queries as one or many updated graph streams. The existing algorithms mainly contribute to single query answer, where queries are set to be isolated and evaluated independently. Currently, the single query answer technique for the dynamic graph is quite mature, as seen in NNT [75], SJ-tree [16], and SSD [30], where algorithms use a-query-at-a-time approaches: optimizing and answering each query separately. They do not allow batch processing of multiple queries. However, sequential processing is not always most efficient when multiple queries arrive [62].
Zervakis et al. [83] first proposes a continuous multi-query process engine, TRIC, over the dynamic graph. To make full use of the information from multiple queries, it decomposes them into several covering paths and constructs an index to organize the paths. Whenever an update occurs, it continuously evaluates queries by taking advantage of the shared limitations in the query set. The key point of multiple queries is to (1) quickly detect the affected queries for each update and (2) avoid expensive joins and explore an approach for larger sets of queries.
In summary, each of the algorithms and classifications presented is different and solves different graph matching problems. Table 2 summarizes the representative continuous subgraph matching algorithms and their classifications. The following sections will detail the continuous subgraph matching technique based on structure- and content-based change. In each section, the main problems will be analyzed and compared, and representative algorithms and research status of each matching technique type will be discussed. Since most current research revolves around the problem of continuous subgraph matching for structure-based change, the main focus of this paper will be on the latest research progress on graph matching techniques for structure-based change.
3 Continuous subgraph matching for structure-based change
Continuous subgraph matching for structure-based changes is the most widely used technique currently. From the perspective of algorithm design, two techniques currently exist snapshot-based matching (Sect. 3.1) and incremental matching (Sect. 3.2). The snapshot-based matching technique treats the updated data graph for each timestamp as a static graph to perform a matching algorithm. In contrast, the incremental matching technique only analyzes and matches the updated parts of the data graph, avoiding the unnecessary calculation of rematching the overall data graph. Furthermore, the finer-grained classification of the incremental matching technique is described in detail in Sect. 3.2. In Fig. 5, we summarize the classification and representative algorithms of the continuous subgraph matching technique for structural change.
3.1 Snapshot-based matching technique
Snapshot-based matching technique over dynamic graph is regarded as a large-scale static graph matching problem since each updated data graph for each moment is a static graph. Matching algorithms are then performed on these continuous static graph streams. It is usually suitable the cases where there are numerous added edges. In this case, adding all the edges can be completed simultaneously, and the matching algorithm is performed on the updated data graph snapshot. Therefore, the snapshot-based matching technique includes two parts, an update operation and a matching operation. The following is a discussion of these two operations.
3.1.1 Data graph update
The continuous subgraph matching problem was first discussed in [75] by Wang et al. In this study, continuously updated graph data was regarded as graph streams, and a snapshot-based pattern matching algorithm, the NNT algorithm, is proposed, which has achieved good performance. The NNT algorithm constructed a node-neighbor tree(NNT) for each vertex u in both data graph G and pattern graph P, which is denoted as NNT(u). For the data graph G, given a depth value L, \(NNT(u) (u \in G)\) stores all the paths in the data graph G, which regards u as the root vertex, and the length does not exceed L. As shown in Fig. 5a, \(T_1, T_2, T_3,\) and \(T_4\) are NNTs with a depth value \(L=2\) corresponding to vertices 1, 2, 3, and 4 in the data graph G. The capital letters indicating the labels of the vertices, the contents of \(T_1\) and \(T_2\) are the same. Figure 6b shows an inverted index for NNTs to find the vertices and edges in each NNT that match the vertices and edges in the data graph, where \(*\) represents the omitted part in the inverted index.
The update operations for structure changes over dynamic graph include deleting edge operations and adding edge operations. The following discussions show how each operation uses the inverted index based on the NNT to update the index.
Delete edge operation: Assuming that an edge in a data graph is deleted at a certain time, the corresponding edges and sub-edges in all NNTs have to be deleted simultaneously through the inverted index. For the data graph G in Fig. 5, if edge (1, 3) is deleted, edge (a, c), sub-edges (c, e) and (c, f), and the vertices associated with these edges \( T_1 \& T_2\) are removed from the inverted index.
Add edge operation: If an edge is added to a data graph at a certain time. First, the vertices in each NNT, corresponding to the node of the added edge, are found based on the inverted index. Then, the newly generated path in the data graph is added to the NNT, starting from the vertex of the corresponding node. For the data graph G in Fig. 6, if edge (1, 4) is added, in \(T_1\), the path \((a \rightarrow g \rightarrow h)\) is added from vertex a, where the attributes of vertices g and h are C and B, respectively. In \(T_2\), the path \((b \rightarrow g)\) is added from vertex b, where the attribute of g is C. The adding edge operation is completed when all paths in the NNTs are added. Finally, the inverted index needs to be updated by adding the newly added vertices and edges. Similarly, the same operation needs to be performed on vertex 4 to update the index structure.
The NNT algorithm matches the pattern graph with the snapshot of the data graph at each moment, which does not reflect the evolution process of the graph over time. Yang et al. [81] first proposed an index structure, BR-Index, for large-scale dynamic graphs, which divided the data graph into a series of overlapping index regions. And each region contained several independent core regions. It extracted the maximum features (small subgraphs) of each index region and then established a feature lattice for maintenance and lookup. Meanwhile, it also maintained a vertex lookup table based on the hash principle. The table was used to store the data vertices in each index region and which core region the vertex is in the index region. Thus, the updated part can be located quickly using the lookup table when a data graph is updated.
Additionally, Song et al. [66] proposed a subgraph matching algorithm, DDST, based on a time window, which was the first work to impose a timing order constraint on graph streams, where the window contained all the snapshots that met the time constraint. Moreover, there was a time constant on each edge that recorded the generation time of the edge. Based on this timestamp, it was possible to determine whether a legitimate data subgraph is within a given time window. Only when the legal data subgraph in the window met the matching conditions, it would become a matching result. Compared with the NNT algorithm, the advantage of a time window approach is its ability to evolve the graph over time and that it conforms better to practical applications.
Furthermore, Khurana et al. [42] proposed a tree-like index structure, DeltaGraph. It is a rooted and directed graph where each leaf node stores a snapshot arranged in time order, and the internal nodes store a graph made up of combining lower-level graphs. Here, the root node represented the source node, the leaf node represented the target node, and each edge held information from the source and target nodes. When the data graph performed an add or delete edge operation, this index structure added a snapshot of the data graph at that moment to the leaf node. The information stored with each edge is called edge delta. And it is sufficient to build the source node (i.e., root node) of the graph corresponding to the target node (e.g., leaf node) of the graph, so that a specific snapshot can be created from the root to the snapshot by traversal the path. When the data graph performed an add or delete edge operation, this index structure only needed to add the snapshot of the data graph at that moment to the leaf node.
3.1.2 Subgraph matching
When using the subgraph isomorphism method in the matching process, the time complexity is very high. For example, for the NNT algorithm using the subgraph isomorphism method for matching, the time complexity can be described as \(O(n_1 *n_2(|T_1|^{1.5} / log|T_1|)|T_2|)\) [81], where \(n_1\) represents the number of vertices in the pattern graph, \(n_2\) represents the number of vertices in the data graph at each time, \(T_1\) and \(T_2\) represent the maximum number of NNTs corresponding to the pattern graph and data graph. This shows that the subgraph isomorphism method severely restricts the execution efficiency of the NNT algorithm; therefore, it does not meet the real-time requirements of continuous subgraph matching and should be avoided as much as possible.
NNT algorithm uses an approximate algorithm, which performs subgraph matching by comparing the paths of two vertices in the NNT. If all the paths in the NNT constructed by each pattern graph vertex can find a match in the NNT constructed by the corresponding data graph vertex, it indicates that the pattern graph matches the data subgraph. The time complexity of this subgraph matching method is \(O(n_1n_2r^l)\), where r represents the maximum degree of the data graph node, and l represents the selected depth value. Because each matching operation requires matching all possible vertices, the overhead is still very high. Thus, the algorithm also proposes a coding method to convert the NNT into a numerical vector and counts the number of different paths in all NNTs to further reduce the computing overhead.
As shown in Fig. 6a, there are eight different paths in \(T_1, T_2, T_3\), and \(T_4\); hence, the NNT of each vertex can be represented by an 8-dimensional vector, where the i-th dimension records the number of i-th paths in the NNT. Consequently, the pattern graph and subgraph can be converted into a multidimensional numerical vector. If the value of each dimension in the pattern graph is less than or equal to the value of the dimension in the subgraph, it means the subgraph meets the matching conditions. The matching time complexity based on the above optimization is \(O({{\overline{L}}}n_1n_2)\), where L represents the number of non-zero items in the numerical vector.
During the subgraph matching process, the BR-Index algorithm first extracts the features (subgraphs) set of the pattern graph. Then based on this extracted set, it finds the index regions in the data graph that contain some of these features. Next, it finds the candidate sets of these features in the corresponding index region. Finally, the candidate sets are combined to obtain the matching result.
In comparison, the DDST algorithm uses a simulation matching method to complete subgraph matching. As simulation matching is more flexible than isomorphism matching, it can identify more useful results. However, in subgraph isomorphism traditional graph simulation extends edge-to-edge mapping to edge-to-path mapping and imposes a more flexible topological constraint, resulting in imprecise results. To solve this problem, the DDST algorithm adds two constraints Dual simulation and Locality [50] to graph simulation and constructs a signature for the pattern graph. The signature includes the labels of the edges and vertices, as well as the in- and out-degree information of all vertices. If the signature of the data subgraph is consistent with the pattern graph, it sequentially uses the binary simulation. Simultaneously, it presents the time order of the edges in the pattern graph. If the time attribute of each edge in the data subgraph meets the time order in the pattern graph, it meets the matching condition. Further, DeltaGraph uses a distributed parallel graph processing framework to complete subgraph matching; it stores snapshots in memory. Given a pattern graph, it can directly find a matched snapshot starting from the root using the information on the edge.
In summary, the subgraph isomorphism method has a low computation efficiency and is very time-consuming. Therefore, the approximate matching algorithm and simulation matching are better candidates for the design of snapshot-based matching methods. The approximate algorithm uses a small number of false-positive results to reduce the matching time, and the simulation matching uses the verification of binary relationships to replace the subgraph isomorphism. Simulation matching is mainly applied for applications that focus more on mining relationships between vertices. Finally, the application requirements determine the most suitable subgraph matching algorithm.
3.2 Incremental matching technique
Using the snapshot-based matching method over dynamic graphs will cause many redundant calculations even though there is only a small number of update operations. In response, Wenfei Fan et al. [23] first applied the incremental processing technique to continuous subgraph matching by proposing the IncSimMatch algorithm and IncBMatch algorithm, which only analyzed and matched the updated parts of the data graph. In addition, they compared the two algorithms with their batch processing (snapshot) algorithms, Match\(_{s}\) and Match\(_{bs}\) , respectively. The experimental results showed that when the number of addition or deletion edge operations did not exceed a certain percentage of the total number of edges in the initial data graph, the execution efficiency of the incremental matching technique was higher.
Incremental matching means that given a data graph G, a pattern graph P, an initial pattern graph matching result P(G), and a set of update operations for data graph \(\Delta G\), a newly added matching result set \(\Delta O\) will be calculated after the data graph update. This is \(P(G \oplus \Delta G) = P(G) \oplus \Delta O\), where \(\oplus \) represents the operator used to add the changed content to the original data.
In the matching process, the incremental matching technique can either use direct computation approach or construct an auxiliary data structure to utilize the intermediate results. The advantages and disadvantages of the two methods are discussed in detail below.
3.2.1 Direct computation
When an edge is added or deleted, it may only affect a small part of the subgraph. Therefore, Wenfei Fan et al. [23] proposed an incremental algorithm, IncIsoMat, based on direct computation with the graph simulation method. It identifies and extracts a subgraph \(G'\), which can be affected by \(\Delta O_i\). It executes a subgraph matching method on \(G'\) to obtain the change to the original matches by determining the set difference between the original matches and new matches. Wenfei Fan et al. [22] further proposed another direct computation method, IncISO, for the incremental subgraph isomorphism problem. When the data graph is updated, only the data graph nodes within the diameter range of the pattern graph around the updated edge are rematched to avoid double calculation of the entire data graph. The cost of IncISO can be represented as a function of |P| and \(|G_{d_P}(\Delta G)|\), instead of the entire graph size |G|, where \(G_d(v)\) represents the \(d-\)neighbor subgraph of v that are within d hops.
Although the above algorithms reduce redundant calculations by matching subgraphs in the affected small-scale subgraphs, they also create new performance problems. These approaches based on repeated search may incur significant overhead in extracting the affected subgraph \(G'\) (resp. \(G_d(v)\)), performing subgraph matching on \(G'\) (resp. \(G_d(v)\)) and computing the newer matching for each \(\Delta O_i\). Moreover, due to the average distance between two vertices is extremely small in real-world graphs [71], the extracted subgraph \(G'\) (resp. \(G_d(v)\)) could include most of the nodes and edges in G, leading to a useless optimization.
In terms of graph databases that handle continuous subgraph matching, many existing graph databases, such as Neo4j [3] and OrientDB [4], only support one-time subgraph queries which evaluate the subgraph on each snapshot of the data graph. However, due to the real-time demand, many applications are required to handle continuous subgraph queries. To solve this problem, Kankanamge et al. [40] presented an active graph database, Graphflow, which supports incremental subgraph evaluating for each update. Internally, the system’s query processor is based on Generic Join [58], which is essentially a worst-case optimal join algorithm [57, 73]. In addition, Ammar et al. [10] further extend the worst-case optimal join to the distributed system.
Given a pattern graph \(P=(V(P), E(P))\), the generic join first determines a query node order \(s = \langle u_1, u_2, \ldots ,u_{|E(P)|}\rangle \), and then, implements a multi-way join using the multi-way intersection operation on the query nodes in turn, according to order s. Each order of the query node can also be considered as a query plan. In the problem of subgraph matching, the multi-way intersection can be achieved using the intersection operation on adjacency lists matched by one or more query nodes. In the process of implementation, the order, s, of query nodes ensures that, for any k less than |E(Q)|, the induced subgraph composed by \(s = \langle u_1, u_2, \ldots ,u_k \rangle \) is connected. Here, the induced subgraph is represented as \(P_k\). The entire process of a generic join contains two basic operations:
-
Scan: For first two nodes on the query node, order \(s = \langle u_1, u_2, \ldots ,u_{|E(P)|}\rangle \) is obtained directly by scanning the adjacency list of the graph.
-
Extend/Intersect: When obtaining the \(P_{k-1}\) match, the matches of \(P_k\) can be calculated through the Extend/Intersect operator. For any match of \(P_{k-1}\) , each match of the query nodes adjacent to \(u_k\) starts along its adjacency table to acquire all possible matches for \(u_k\); then, these matches are intersected to obtain the final matches for \(u_k\).
Although Graphflow can handle the continuous subgraph query based on Generic Join without calculating the set difference, it still needs to compute the join operation from scratch for each \(\Delta O_i\) even if the \(\Delta O_i\) does not cause any new matches.
3.2.2 Auxiliary data structure
Although the incremental matching algorithm with direct computation is performed on a small-scale graph affected by \(\Delta O_i\), it still needs to perform subgraph matching on the small graph from scratch for each update, leading to high calculation costs. To solve this problem, an auxiliary data structure can be constructed to keep track of the (partial) results of the previous computation. Then, for each recent update, the newer results can be obtained easily.
In the process of constructing an auxiliary data structure, the intermediate results can either be stored in the query graph or data graph, which is called query-centric representation or data-centric representation, respectively. After the auxiliary data structure is constructed and the candidate set of all query vertices is found, the subgraph matching operation of the query graph is performed. The following is an in-depth introduction to subgraph matching based on the two different auxiliary data structures.
Query-Centric Representation Query-centric representation is the more commonly used, which stores a set of candidate data vertices for each query vertex, and partial matches can be obtained by traversing the query graph. For the subgraph matching process, continuous subgraph matching is similar to static subgraph matching. Based on whether a pattern graph needs to be decomposed, it can use either a join-based matching or an exploration-based matching technique. Further, graph simulation-based matching technique has also obtained good experimental results in continuous subgraph matching. An in-depth introduction to these three methods is provided below.
(1) Join-based matching technique
For a large pattern graph, it can be time-consuming to rematch the entire pattern graph every time the data graph is updated. Therefore, rather than looking for a match to any edge of the entire graph or the query graph, it is better to divide the query graph into smaller subgraphs and then search for them. As shown in Fig. 7, the pattern graph P is decomposed into a series of smaller pattern subgraphs, expressed as \(\langle P_1, \ldots ,P_k\rangle \). These pattern subgraphs have then tracked the matches with individual subgraphs and combined the matches to produce progressively final matching results for the entire pattern graph.
The join-based method is widely used in static subgraph matching. The representative algorithms include the gIndex algorithm [80] and GraphGrep algorithm [31]. The join-based method mines features using recognition capabilities from the data graph, such as path [31, 87], tree [63, 82] and subgraph [15, 37, 80]. Based on these features, it can build an index on the data graph and decompose the pattern graph. However, simply applying this method to the continuous subgraph matching process will produce a series of problems. For example, the gIndex algorithm [80] would need to mine the features of the data graph subgraph at each moment. Therefore, this method is unsuitable for continuous subgraph matching processing with real-time analysis requirements. Although the GraphGrep algorithm [31] meets the real-time requirements, it may produce several negative results by only using the path; therefore, it is not highly recommended for verification and filtering applications. Notably, when applying the join-based method to continuous subgraph matching analysis, the efficient extraction of the data graph features is the main challenge.
Data graphs constructed in social network applications are often multi-relational graphs. The properties of edges represent the connectivity and relationship between entities. Therefore, in this application, the relationship between data graph entities (i.e., type of edge) can be used as a data graph feature to decompose the pattern graph. Choudhury et al. [16] proposed a tree-like auxiliary data structure called the subgraph join tree (SJ-Tree). The SJ-Tree is a binary tree whose root node represents the pattern graph, and its child node represents the subgraph of the pattern graph. And then, the subgraph of each node is further decomposed to obtain their child node. The leaf nodes represent the final decomposition results.
Decomposition of social query in SJ-Tree (Taken from [16])
As shown in Fig. 8, each node in the SJ-Tree stores the information of its sibling and parent nodes and maintains a hash table to store the matching results of the node. When the data graph is updated, an iterative search of all the leaf nodes on the SJ-Tree is conducted to obtain the matching result that contains the newly added edge; this matching result is stored in the hash table of the corresponding node in the SJ-Tree. Simultaneously, where possible, the matching result of the leaf nodes is integrated with the matching result of its sibling nodes to form a larger matching result. The larger matching result is stored in the hash table of the parent node. Furthermore, a join order is defined, where the individual matching subgraphs will be combined. The join operation is complete when a result that matches the entire pattern graph is generated. Figure 8 shows an example of a social query decomposition with SJ-Tree. The leaf nodes candidate set of SJ-Tree {(“George”, “friend”, “Join”)} and {(“Join”, “like”, “Santana”)} can be connected and integrated to be a greater matching result {(“George”, “friend”, “Join”), (“Join”, “like”, “Santana”)}.
Although SJ-Tree can perform graph queries in an exponential time relative to its height, it still has a limitation. In Fig. 8, since the “friend" relationship frequently occurs in the data graph during the subgraph matching process, it will undoubtedly take a considerable amount of time to track all the edges matching “friend". Therefore, matching the “friend" edge in the pattern graph can be postponed, and the decomposition result with relatively infrequent occurrences in the data graph is matched first. Based on this idea, Choudhury et al. [18] proposed a Lazy-search algorithm. Given an initial data graph G, the selectivity of a k-edge subgraph g in graph G is the ratio of the number of occurrences of g to the number of all k-edge subgraphs in G, where g is called the selectivity primitive. To both limit the computational cost of subgraph isomorphism and keep the selectivity primitive effective when a data graph updates, unilateral or bilateral subgraphs are generally selected as the selectivity primitive. The Lazy-search algorithm calculates the selectivity of the types of unilateral or bilateral subgraphs contained in the data graph with an offline method and sorts these selectivity primitives in ascending order according to their selectivity value. When the selectivity value is lower, the recognition ability is higher. Subsequently, the query graph is decomposed according to the order of these selectivity primitives.
In the matching process, in order to ensure that the matching results of adjacent leaf nodes in the SJ-Tree are still adjacent on the data graph, it constructs a bitmap index, as shown in Fig. 9b. The rows represent all the nodes in the data graph, and the columns represent the decomposition results of a pattern graph that satisfies the adjacency relationship. Figure 9a shows the two adjacent decomposition results of the pattern graph P. If the data graph vertices are in the matching result of the query fragment \(g_1\), the corresponding bits are 1, and the others are 0. For example, as \(g_1\) matches the edge \((u_1, u_2)\) in the data graph, the bits of \(u_1\) and \(u_2\) corresponding to \(g_1\) are 1, and the other bit positions are 0. When matching \(g_2\), only the results that meet the matching condition of \(g_2\) around the vertex for which the bitmap vector is 1 in \(g_1\) need to be found. This advantage is that it can use adjacent decomposition results to ensure that the matching results are still adjacent on the data graph. Therefore, it can avoid matching results that do not meet the adjacency relation, reduce the search space, and speed up the matching process.
Based on the decomposition and join methods, Youhuan Li et al. [48] studied continuous subgraph matching with timing order constraints which meant the occurrence of edges in the stream followed the time sequence, and proposed the timing subgraph matching algorithm. All the matching results should meet both structure and timing constraints. An example of query graph Q with two-timing order constraints is presented in Fig. 10, \(\epsilon _1\) \(\prec \) \(\epsilon _2\) indicate that edges matching \(\epsilon _1\) should arrive before edges matching \(\epsilon _2\) in the subgraph matches of Q. In Fig. 10, the query is decomposed into a set of subgraph queries that contain only one timing order constraint called timing connect-query, and a match-store tree (MS-tree) for each subgraph query \(Q_i\) is constructed to store the intermediate results \(G(Q_i)\). Then, all the intermediate results of the subgraphs are joined according to the timing order constraint to obtain the final matching result G(Q)=\(\{G(Q_1),G(Q_2),G(Q_3)\}\).
Based on timed order constraints, MS-Tree can filter out a large number of discardable partial matches, which reduces both space costs and maintenance overhead without incurring any additional data access burden. Unlike the DDST algorithm with the time constraint mentioned earlier, this is based on subgraph isomorphism rather than graph simulation. More importantly, it was designed to perform effective concurrency management, using a fine-granularity locking technique in the computation to improve system throughput. It is the first work that studied subgraph matching with concurrency management over streaming graphs.
Join-based matching techniques use subgraph isomorphism in the matching process, which can obtain accurate matching results, but at an expense. Although the method of query decomposition can control the size of subgraphs and limit the cost of the subgraph isomorphism, it still produces many invalid intermediate results. If an index is built for the data graph to reduce the number of invalid intermediate results, the cost of index construction and maintenance should also be considered. Simultaneously, how to mine feature structures over frequently updated dynamic graph data for query decomposition is still a problem that needs to be solved.
(2) Exploration-based matching technique
Motivated by the above problems, Sun et al. [69] proposed an exploration-based matching technique, where the obtained results are approximate rather than exact. An example of the exploration process is shown in Fig. 11. Given a pattern graph P and a data graph G, this technique starts with vertex a in the pattern graph and finds the matching vertex \(a_1\) in the data graph G through a simple index that maps labels to node IDs. Next, the data graph is explored from vertex \(a_1\) to reach vertex \(b_1\), meeting the requirement of the partial query (a, b). Then, it is explored from \(b_1\) to reach \(c_1\) and \(c_2\), meeting the requirement of the partial query (b, c). In this way, it can obtain the matching results of the pattern graph from the data graph without generating and joining large intermediary results or building and maintaining the index for the data graph like the join-based matching technique does. Certainly, if the approach starts from a node labeled b or c, it may still get some useless intermediate results. But in general, it will not generate as many of them unless it is in the worst case. The meaningful advantage of the exploration-based matching technique is that the join operations are avoided.
Compared with the join-based continuous subgraph matching, the explorat-ion-based continuous subgraph matching is more suitable for the distributed parallel graph processing framework. The mainstream distributed parallel graph processing frameworks, such as Google’s Pregel [51] system, use a node-centric computing model framework. For this framework, each vertex maintains an input queue and an output queue. Additionally, each computing task is composed of a series of super-steps. After receiving the input information from the input queue in each super-step, the vertex will process the information according to the user-defined script program, and finally, output the processing result to the output queue. As the storage and processing units are vertices, this computing model, together with join-based continuous subgraph matching, will produce several intermediate results, thereby increasing the processing, storage, and data transmission costs of vertices. In summary, the join-based continuous subgraph matching technique is unsuitable in a distributed parallel graph processing framework. In comparison, the exploration-based method is more suitable because it does not require index construction, nor does it produce and process numerous intermediate results. Furthermore, it naturally meets the potential needs of incremental computing.
The PathMatch algorithm is an exploration-based matching technique used in a distributed parallel graph processing framework. It finds a Hamiltonian path in the pattern graph that can access each vertex in the graph. Using this path transfers information to complete the exploration (matching). However, if we apply the exploration-based matching technique to a distributed parallel graph processing framework, it still faces the following problems:
-
First of all, the matching results obtained by the exploration-based method are approximate rather than exact;
-
When solving some continuous subgraph matching problems, it is more expensive to use a naive graph exploration than the join operation;
-
Not all queries can be answered by relying only on exploration, for example, when attempting to check whether the next explored vertex is the first matching vertex.
To solve the above problems, Sun et al. [69] presented a novel exploration method, STwigMatch, that maximizes the benefits of both the join-based approach and the exploration-based approach while avoiding their disadvantages. Specifically, this method uses a join-based graph matching framework on the macro level and an exploration-based matching method to avoid useless candidates during the join and find matching results. In other words, it decomposes the pattern graph into a series of small twigs based on the transmitted information in the pattern graph rather than the feature subgraph extracted in the data graph and then stores each decomposition result on a vertex of the distributed parallel graph processing framework. Later, it matches these small twigs through the exploration-based approach. The benefit of this algorithm is the tradeoff between matching calculation cost and matching result accuracy.
Additionally, Gao et al. [30] proposed an exploration-based algorithm, SSD, in the distributed framework, Giraph. Figure 12 is an illustration of the computation process of the SSD algorithm. First, a vertex with the maximum degree is labeled as the sink vertex in the pattern graph (i.e., vertex C in Fig. 12a). It is called a sink, as all the edges satisfied are passed to it, and it does not need to send a message out. After selecting the sink vertex, the direction of the edge is determined via a BFS (breadth-first-searching) strategy. Based on this concept, the pattern graph can be converted into a directed acyclic graph (DAG) with a single sink vertex as in Fig. 12b. And regarding the sink vertex as a cut vertex, the DAG can further be decomposed into three sub-DAGs. In each sub-DAG, there are some source vertices (e.g., vertex a and e in sub-DAG 0), which do not have any incoming edge and can initialize the message transfer.
Figure 12c shows the process of subgraph matching. In sub-DAG 0, information passes to all its neighbors (vertex 1 and vertex 2) to convert from vertex 0. Then, the converted information continues to pass to their downstream neighbors and finally reaches vertex 3. Thus, it can obtain the information transmission rules from the source vertex. Then, it maps the information transmission rules to the data graph and starts from the vertex in the data graph with the same attribute as the source vertex to check whether the subsequent vertices match the query vertices based on the rules. For example, in Fig. 12c, source vertex 0 matches the data graph vertex a, and the matching process is completed by a series of super-steps. In super-step 1, vertex a passes information to its neighbors; in super-step 2, vertices b and c pass the information to vertex d after receiving it. If vertex d receives two pieces of information from the same vertex, which is the same as the information transmission rules of sub-DAG 0, the existence of a matching subgraph can be proved.
Illustration of the SSD algorithm (Taken from [30])
However, there is still a problem with the SSD algorithm. In super-step 2, if edge (a, b) is deleted when vertex b and c pass the information to vertex d, vertex d will return an expired, inaccurate matching result after receiving the information. The problem of inconsistent results is severe in distributed systems since each distributed parallel graph computing framework contains many super-steps. In order to solve this problem, Gao et al. [29] proposed the Stp(Q) algorithm.
The exploration-based matching method is more suitable for processing large-scale graphs; however, inconsistent matching can produce inaccurate matching results. Therefore, this type of method is suitable for applications that do not have strict accuracy requirements for results, such as social network analysis. Determining methods to effectively combine the exploration-based and join-based matching methods to balance efficiency and subgraph matching accuracy is a topical issue in current research.
(3) Simulation-based matching technique
Note that the exploration-based matching technique produces inaccurate matching results, while the join-based matching technique is more expensive. Both methods use the NP-hard subgraph isomorphism matching method, which limits the improvement of the performance. Therefore, the simulation-based matching technique has gained attention in the continuous subgraph matching field. This method generates a matching candidate set for each query vertex according to its label and then filters out the unmatched vertices according to the different approximation degrees of the precursors and successors of the query vertices (see Sect. 2.1 for details on simulation matching).
Wenfei Fan et al. [23] proposed an incremental graph simulation matching (IncSimMatch) algorithm with the auxiliary data structures, match(v) and candt(v), to maintain the intermediate results and accelerate the matching calculation. Here, v represents a vertex in the pattern graph, match(v) represents a vertex in the data graph that matches v, and candt(v) represents a vertex in the data graph that has the same label as that of v but does not meet other matching conditions.
With these auxiliary data structures, unnecessary update operations can be filtered out:
-
For delete edge operation, only when the deleted edge, such as \((u_i, u_j)\), satisfies \(u_i \in match(\cdot )\) and \(u_j \in match(\cdot )\), it will cause the reduction of the matching results.
-
For add edge operation, only when the added edge, such as \((u_i, u_j)\), satisfies \(u_i \in candt(\cdot )\) and \(u_j \in match(\cdot )\) or \(u_i \in candt(\cdot )\) and \(u_j \in candt(\cdot )\), it will cause a new matching result set.
In addition, Wenfei Fan et al. extended the definition of initial simulation matching and proposed the concept of bounded simulation, which redefines the pattern graph with edge weights. Each edge in the pattern graph maintains a constant, k. If there is a vertex in the data graph that matches the successor vertex of the corresponding vertex in the pattern graph within k hops, then there is a match. The author further applied the bounded simulation matching technique to the continuous subgraph matching and proposed the incremental algorithm, IncBMatch. The IncBMatch algorithm is similar to the IncSimMatch algorithm mentioned with the auxiliary data structure match(v) and candt(v), which solves the problem of highly complex isomorphism matching and achieves good results. The advantage of bounded simulation matching is that the matching process can be completed in polynomial time, which effectively improves the efficiency of subgraph matching.
The literature [41] was the first work that applied the IncSimMatch algorithm to the vertex-centric distributed parallel graph processing framework and realized the distributed parallel computing of dynamic graph simulation matching. During the matching process, the updated graph was stored in a vertex of the framework, and an execution script allowed the vertex to filter out edges that would not produce new matching results. Then, the edges were evaluated against the edge constraint relationship in the graph simulation matching by a vertex of the framework. The main process would receive the processing information from all vertices and then evaluate whether these subgraphs meet the edge constraint relationship in the graph simulation matching to obtain the final matching result.
However, the simulation matching method is based on a binary relationship and can produce inconsistent results with the structure of the pattern graph; therefore, it is mainly suitable for applications that do not have very strict requirements for graph structure matching, such as social network analysis. Further, simulation matching can be used to prune candidate sets. Wickramaarachchi et al. [76] proposed a distributed pruning algorithm D-IDS over the dynamic graph, which uses a dual simulation matching method to prune the data graph. The dual simulation requires that all child and parent nodes of the current node conform to the binary relationship. When the data graph is updated, the binary simulation is used to prune the data graph, and a large data graph can be pruned into a relatively small data graph. Meanwhile, the data graph can be maintained continuously. In the matching process, only incremental matching needs to be performed on the small graph.
The matching efficiency of simulation matching is high; it is applicable to both distributed and centralized environments and has some unique advantages. This advantage is more apparent in the processing of continuous subgraph matching. The bidirectional simulation matching based on graph simulation matching can better tradeoff the effectiveness and timeliness of simulation matching results and simultaneously obtain a matching result that is more consistent with the structure of the pattern graph, thereby effectively compensating for the disadvantages of the join-based matching technique and exploration-based matching technique.
Although constructing an auxiliary data structure to store intermediate results can reduce the re-computation overhead and achieve better performance, query-centric representation still has several limitations. When a data graph vertex v has multiple candidate query vertices, v needs to be copied as many times as the number of the matched query vertices. Further, storing many partial results and then joining each other follows a certain order does cause lots of redundancy. Specifically, the worst storage complexity for SJ-tree is \(O(|V(q)|*|E(G)|^{|E(q)|})\), where the |E(G)| represents the number of edge in a graph G, and V(q) represents the number of vertices in a graph q. Finally, when any update occurs for vertex v, it must find all the partial matches containing v in the query-centric representation. Therefore, it needs to design and maintain an additional index of duplicate keys to find corresponding partial matches in the query-centric representation.
Data-Centric Representation All of the above problems of existing methods motivated [43] to investigate the novel concept representation, called data-centric graph (DCG). It stores the corresponding query vertex ID as the incoming edge label for each data vertex in the data graph. Thus, in the DCG, each data vertex appears at most once, and each data edge stores at most |V(q)| edges. Thus, its worst-case storage complexity is \(O(|V(q)|*|E(g)|)\). For a dynamic graph, the execution model allows for fast incremental maintenance. Each edge in the DCG has one of three states: NULL, IMPLICIT, or EXPLICIT. An explicit edge \((v,u',v')\) represents that query vertex \(u'\) is candidate of \(v'\) and the data path and subtree of \(u'\) matches the corresponding data path and subtree of \(v'\). In an implicit edge \((v,u',v')\), only the data path of \(u'\) matches the corresponding data path of \(v'\), and the subtree does not match. When a new/expired edge is inserted/deleted, TurboFlux uses an edge transition model to change the state of each corresponding edge, and finally, it can report positive/negative matches based on the explicit edge. Furthermore, compared with other auxiliary data structures, as DCG itself is a graph, whenever the DCG is updated, it can directly access corresponding vertices; there is no need for an additional duplicate key index like with query-centric representation.
A running example (taken from [43])
Figure 13 shows a running example of DCG. Figure 13 shows the DCG obtained after the transformation of the data graph for the query graph q. When \((v_0,v_1)\) is inserted into g, the state of \((v_0,u_1,v_1)\) in the DCG will be transited from NULL to IMPLICIT, as the inserted edge matches \((u_0, u_1)\) and \(v_0\) has an incoming explicit edge with label \(u_0\) (Fig. 13). Then, the state of \((v_1,u_4,v_4)\) is transited from NULL to IMPLICIT, because \(v_4\) is the child vertex of \(v_1\) and the state of \((v_0,u_1,v_1)\) is IMPLICIT (Fig. 13e). Next, the state of edge \((v_1,u_4,v_4)\) will be transited to EXPLICIT (Fig. 13f). Note that the subtree of \(v_1\) matches that of \(u_1\). Thus, the state of \((v_0,u_1,v_1)\) is transited to EXPLICIT (Fig. 13g). Finally, the state of \((v^{*}_s,u_0,v_0)\) is also transited from IMPLICIT to EXPLICIT for the same reason (Fig. 13h).
In summary, the concise auxiliary data structure and efficient incremental maintenance strategy of TurboFlux make it the most advanced method currently. It can efficiently identify update operations that may cause positive/negative matches with the current partial solutions.
3.2.3 Complexity analysis
Continuous subgraph matching algorithms aim to find the occurrences of a given pattern on a stream of data graphs online. The dynamic nature of the data graph demands that the matching results need to be responded to in a little time. Thus, the incremental continuous subgraph matching algorithms have been widely studied. In Table 3, we summarize the space complexity of the index and the time complexity of updating the index on each update of four latest and representative continuous subgraph matching algorithms, including IncIsoMatch, SJ-Tree, Graphflow, and TurboFlux. IncIsoMatch and Graphflow are the direct computation incremental algorithm, and they do not generate any auxiliary data structure. While SJ-Tree and TurboFlux construct an auxiliary data structure to accelerate the filtration rate.
From Table 3, we can see that for SJ-Tree, the space complexity and time complexity of the auxiliary data structure (index) are exponential, while for TurboFlux, they are linear. This is because SJ-Tree built a tree index and uses a lot of join operations. TurboFlux used a data-centric representation to store the candidate set. Obviously, TurboFlux can effectively reduce the search space.
4 Continuous subgraph matching for content-based change
Besides the structure-based continuous subgraph matching, the content-based continuous subgraph matching techniques have also been studied. For example, using this technique, a server deployment solution that meets user needs can be found in a frequently updated data center network [60]; advertisers can find closely linked groups of people in frequently changing social networks; an HR can construct a team in dynamic social networks based on constrained pattern graph [44]. The labels of nodes or edges in these applications will frequently change with time. Currently, there are few studies on content-based continuous subgraph matching techniques. The following are some typical studies.
4.1 Join-based matching approach
Taking resource allocation in a data center network as the application background, Bo Zong et al. [88] proposed the Gradin algorithm to solve the continuous subgraph matching problem between the user-defined resource pattern graph and data-center network. It adopts a join-based matching approach since the data center network has a relatively stable topology but frequent changes in the node/edge labels, which representing vacant CPU cycles and network bandwidth. The matching process will include the following two stages.
-
Offline index construction: First, the frequent graph structure set is decided by existing structure selection algorithms [80], and then all the graph fragments in data that contain the frequent graph structure were obtained by the subgraph mining technique [79]. Based on these frequent subgraphs, an inverted index is built on the data graph.
-
Online query processing: Gradin uses a join-based approach to search the compatible subgraphs of pattern graphs. The pattern graph is decomposed first based on the frequent subgraphs. Then Gradin searches the candidates for each query fragment to sional vectors and uses indices to efficiently search them. Then, it combines all matched fragments of the query’s subgraphs via join operation to form compatible subgraphs for pattern graphs.
Obviously, the frequent update is a huge challenge that needs to be addressed. Thus, the Gradin algorithm further used a grid-based index, FracFilter, which can be used to construct indices for frequent subgraphs in the graph and avoid redundant comparisons. As shown in Fig. 14, let \(s_1\) represent a frequent subgraph mined from the data graph. Then, all the subgraphs in the data graph with the same structure as \(s_1\) can be converted into two-dimensional vectors based on their node labels and further mapped into the two-dimensional grid indices, i.e., all the black dot in FracFilter. Simultaneously, the subgraph with the same structure as \(s_1\) in the pattern graph is also mapped to the grid index in the same way indicated by a red square. If the subgraphs of the pattern graph match that of the data graph, the labels of corresponding nodes must satisfy the partial order. Therefore, the two-dimensional grid can be divided into three areas according to the position of the red square. The subgraphs of the data graph, corresponding to the black dots that fall in the green shadow area, \(R_1\), will meet the matching requirements with the subgraphs of the pattern graph. Conversely, the subgraphs of the data graph, corresponding to the black dots that fall in the brown shadow area, \(R_3\), do not meet the matching requirements with the subgraphs of the pattern graph. Therefore, it is only necessary to perform matching computation on the subgraphs corresponding to the black dots in the area between the green and brown shadow areas (i.e., \(R_2\)) to obtain the matching result.
In the searching processing, the Gradin algorithm requires matching \(dn_s/{\lambda ^d}\) \([(\lambda +1)/2]^{d-1}\) times on average, where \(\lambda \) is the density of the grid index, d is the dimension of the grid index, and \(n_s\) is the number of pattern graph slices during the offline index construction. When the nodes/edges labels of the data graph are frequently updated, FracFilter has a lower maintenance cost than other index structures. In average, each graph update only requires \(2(1-1/{\lambda ^d})\) operations.
Mapping a frequent subgraph \(s_1\) to the grid index (Taken from [88])
The Gradin algorithm can also be directly extended to the case where the edge labels in the data graph are frequently updated. In addition to using frequent subgraphs mined in the data graph to decompose the pattern graph, current advanced subgraph structures commonly used in academia can also be used to decompose the pattern graph.
4.2 Exploration-based matching approach
However, the join-based approach cannot be directly extended to large dynamic graphs [69], and it is suitable for queries with static or infrequently updated labels. Since the active attributes in dynamic graphs would cause a lot of updates to the join indexes. Thus, Mondal et al. [56] designed an exploration-based query evaluation system, called CASQD, to continuously detect meaningful and up-to-date insights based on subgraph matching over content-changed graphs.
The principle of the exploration-based approach is to select a set of potential pivot nodes and then explore their neighbors to get a match. The key of this method is how to select pivot nodes efficiently and make the best use of additional information about adjacent nodes. Therefore, CASQD used a monitor, explore and trigger-based approach to select pivot nodes and reduce the search space:
-
Monitor: In the monitoring phase, CASQD summarized the activity node and/or its activity neighbors via a set of models. The monitoring policies have two methods to capture the activity information: (1) directly evaluating the node’s activity predicates and tracking the number of its active neighbors in real-time; (2) calculating probabilistic “estimates" of the active nodes by historical information.
-
Explore: In the exploring phase, if there is a match around a node v based on its model and neighbors’ knowledge, it is called a pivot node. The exploration phase searches for a match around the pivot node and finds a possible result. In addition, it further updates the neighborhood knowledge of the pivot node with the information gathered during the exploration phase.
-
Trigger: The trigger phase is used to let the neighbors of a active node know about any significant change and update their knowledge based on it.
Compared with Gradin, the advantage of the design is that there is no need for frequent subgraph mining on the data graph, and the advanced subgraph structures can be integrated into any query language that supports subgraph pattern matching. In addition, CASQD extends simple tree-structured patterns to more complicated structures, e.g., stars, cliques, or bipartite cliques, and designs different exploration algorithms for finding different primitive patterns.
5 Performance comparison of matching algorithms
In this section, we summarize and analyze the performance of representative algorithms under different classification respectively. We mainly focus on analyzing the performance of continuous subgraph matching method based on structural change.
Datasets. There are multiple real datasets and synthetic datasets used in this evaluation cover a wide range of settings, including:
-
MIT datasets: it was obtained from the mobile communication information of the MIT Media Lab, using a subset of the mobile communication information of 97 fixed persons from January 2004 to May 2005 as the experimental dataset; 300 graph snapshots were generated. Each snapshot represents the state of the dynamic graph at a certain moment.
-
synthetic data: it was generated using a graph generation tool.
-
collection of real datasets: including road network traffic data, telephone communication data, patent citation data, and Twitter data. Table 4 gives detailed statistics on these data, including the maximum in/out degrees of the datasets, an average time interval of increasing edges, and the number of nodes and edges.
-
New York Times dataset: it contains 39,523 nodes and 68,682 edges.
-
livejournal: a friendship network of an online community site LiveJournai, with 4,847,571 nodes and 68,993,773 edges.
-
YouTube: has 14,829 nodes and 58,901 edges, where each node represents a video object that recorded the length, type, and other attributes.
-
citation network: has 17,292 nodes and 61,351 edges, where each node represents a paper object that recorded the title, author, and publication year of the paper.
-
LSBench: comprises an initial graph containing 20,988,361 triples and a graph update stream containing 2,332,065 triple insertions.
Query graphs. The common query graphs used in this section are a set of 100 queries for each size and query type by randomly extracting subgraphs from the data graph. There are two types of queries: tree and graph (having cycles). And query graphs can further be divided into two types based on the density: sparse (\(d_{avg}\) \(\le \) 3) and dense (\(d_{avg}\) > 3). Both sparse and dense are graph queries. For each type, the query graphs are generated with |V(Q)| varied from 4 to 12 in an increment of two.
Metrics. For structural changed continuous subgraph matching algorithm, the most common evaluation metrics are the query process time, which is the elapsed time of the online processing given a graph update stream, and the size of the intermediate results, which shows the pruning optimization ability of the algorithm. In addition, in the exploration-based approach comparison, transferred messages and precision are also evaluated. While for content changed continuous subgraph matching algorithm, the filtering time and query processing time are the main evaluation metrics.
5.1 Structure-based change
According to the classification method in Sect. 2.2, the structure-based continuous subgraph matching technique is divided into two types: matching based on snapshot techniques and matching based on incremental techniques. This section will introduce the performance evaluation of these two types of algorithms.
5.1.1 Snapshot-based technique
The performance of all algorithm can be evaluated in terms of effectiveness and efficiency. Wang et al. [75] compared the proposed NNT algorithm with gIndex [80] and GraphGrep [31], using real and synthetic datasets. The real data was obtained from the mobile communication information of the MIT Media Lab [8], using a subset of the mobile communication information of 97 fixed persons from January 2004 to May 2005 as the experimental dataset; 300 graph snapshots were generated. Each snapshot represents the state of the dynamic graph at a certain moment. The synthetic data were generated using a graph generation tool [45].
The experimental results show that in terms of effectiveness (as shown in Fig. 15a), the GraphGrep algorithm used more than 50% of the matched result as candidate sets, resulting in poor pruning ability and low effectiveness. There was no significant difference in effectiveness between the NNT and gIndex algorithms, as the NNT algorithm produced a candidate set below 6%, and the candidate set of the gIndex algorithm was between 6% 10%. In terms of efficiency (as shown in Fig. 15b), the query time of the gIndex algorithm was significantly higher than that of the other two algorithms. This can be attributed to the frequent subgraph mining method used in gIndex. Although the validity of the matching result is high, frequent subgraphs of the data graph need to be mined for each snapshot, which is time-consuming. The NNT and GraphGrep algorithms did not need to mine the subgraph features, making them more efficient.
Compared with the join-based matching technique (gIndex algorithm) used on static graphs, snapshot-based continuous subgraph matching technique can be improved in two ways. The first method is to design an effective index structure, which can efficiently process the update of the data graphs and use the pruning ability of the index to effectively reduce the size of the matching candidate set. The second method is to use an approximation algorithm, sacrificing some accuracy in exchange for higher matching efficiency.
In [66], there are several real datasets of dynamic graphs introduced, including road network traffic data, telephone communication data, patent citation data, and Twitter data. Simultaneously, the proposed DDST algorithm was compared with the NNT algorithm. The NNT algorithm does not use time window constraints and instead uses the approximation algorithm based on the isomorphic matching problem. Note that this led to errors in the matching results. In the comparison, the DDST algorithm considered the time constraint relationship on the updated edge of the data graph and used an exact algorithm based on the simulation matching problem, leading to more accurate results. Because the performance of the algorithm needs to be compared in the same experimental environment, it was necessary to ignore the time constraints and the accuracy of the results in the comparison process; only the efficiency of the algorithm from the perspective of throughput was compared.
As shown in Fig. 16, the experimental results showed that, for the road and phone datasets, the throughput rate of the DDST algorithm was significantly higher than that of the NNT algorithm. Simultaneously, as the range of the window increased, the throughput rate of the NNT algorithm decreased. This is due to the NNT algorithm expending a significant amount of resources on index maintenance which is more sensitive to the increase of data, and adopting a more restrictive isomorphic matching method. In contrast, DDST used the graph simulation method and showed superior performance in terms of throughput (efficiency).
In summary, to meet the needs of real-time applications, the snapshot-based matching technique can be designed by considering the following two perspectives. First, an efficient index structure needs to be designed. This will ensure no excessive index maintenance costs exist in the process of updating the data graph. Furthermore, an approximate algorithm needs to be designed to improve the efficiency. Second, using simulation matching, restrictions need to be added based on graph simulation to better meet the needs of actual applications. Further, an accurate algorithm based on the binary relationship of the simulation matching needs to be designed to improve the matching efficiency of the algorithm. Using either isomorphic matching or simulation matching is based on the specific application background.
5.1.2 Incremental technique
With the emergence of the incremental technique in continuous subgraph matching, an increasing number of studies are focusing on this area. The incremental technique is more suitable for real-time updated graph data, and it can be divided into two categories: directly computing and building auxiliary data structures. Furthermore, the applied subgraph matching algorithm can be divided into three types: join-based matching, exploration-based matching, and graph-based simulation matching. Next, the performance of these three types of continuous subgraph matching algorithms is analyzed and compared. Finally, a comprehensive comparison of the different representative incremental algorithms is made.
(1) Join-based matching technique
Choudhury et al. [16] compared the SJ-Tree and IncIsoMatch algorithms using the real New York Times dataset from August to October 2011. In Fig. 17, the experimental results show that SJ-Tree is faster than IncIsoMatch, and the performance gap between the two algorithms increased as the data graph size increased. This can be attributed to the fact that incremental matching techniques search around each newly arrived edge during the matching process. For IncIsoMatch, when a new edge arrives, it is necessary to find all nodes within the k-hop range around the edge endpoint, where k represents the diameter of the pattern graph. When the data graph is dense, the subgraphs in the k-hop range will accumulate numerous edges, making the search more expensive and the query rate slower. For SJ-Tree using query decomposition, a large matching graph is transformed into a series of smaller matching graphs, making the algorithm more efficient.
While the selective lazy-search method proposed in the [17] can filter the candidate sets based on a selected structure with recognition ability, effectively reducing the search space. The experimental results show that subgraph isomorphism operations account for 95% of total query time in the query process, thus reducing the number of subgraph isomorphisms. This can effectively increase the performance of the algorithm and result in a search speed that is 10\(\sim \)100 times that of the VF2 algorithm [20].
Furthermore, Youhuan Li et al. [48] compared the time efficiency and space efficiency between Timing algorithm, SJ-tree, and three other state-of-art static subgraph matching algorithms which were performed on the affected area window by window, including QuickSI [63], Turbo\(_{ISO}\) [34], and Boost\(_{ISO}\) [61]. Obviously, the Timing algorithm was faster than the other approaches. This is because the Timing algorithm uses the incremental strategy rather than recomputes each snapshot for each time window. Meanwhile, it filters out some discardable partial matches firstly based on the timing order constraint with MS-tree, while SJ-tree needs to enumerate all the partial matches to find expired ones.
The above experimental results show that performing incremental matching will result in low efficiency if the entire pattern graph is matched with the data graph, especially dense data graphs. Therefore, decomposing the pattern graph is an option but may produce many invalid intermediate results. Simultaneously, for the join-based matching technique using a subgraph isomorphism verification method, the invalid intermediate results will lead to a further reduction in matching efficiency. Therefore, by selecting the feature structure for query decomposition, the invalid intermediate results can be filtered out to speed up the matching rate (Fig. 18).
(2) Exploration-based matching technique
Gao et al. [30] compared the SSD with STwigMatch and PathMatch algorithms on the distributed platform Giraph [1]. The study used the real dataset livejournal. STwigMatch used a decomposition framework based on STwig and an exploration-based matching method. The PathMatch method only used the exploration-based matching method. The experiment used query time and total transferred messages as evaluation indices. As shown in Fig. 19, the performance of the SSD algorithm and STwigMatch was better than PathMatch overall, and the SSD algorithm needed to transfer the least information. PathMatch and STwigMatch transfer larger amounts of information, which is time-consuming; the SSD algorithm uses DAG decomposition, which can effectively reduce the information delivery and query response time. In addition, for pattern graphs with a size larger than 7, the memory limit was exceeded on both STwigMatch and PathMatch algorithms, leading to a slow query process.
Gao et al. [29] further proposed the Stp(Q) algorithm based on SSD, which solved the inconsistency of results caused by the super-steps of the SSD algorithm during the process of data graph changes. For Fig. 19, the UpdateRation represents the update ratio of the data graph. The experimental results show that the accuracy of the SSD algorithm decreased with an increase in the degree of change in the data graph or pattern graph. Meanwhile, the SSD algorithm using Stp(Q) could effectively guarantee an accuracy rate of 100%.
The preceding discussions show that using exploration-based matching alone results in poor performance. Therefore, a combination method with join-based and exploration-based approaches could be designed to improve the performance, decomposing the query framework using the join-based method, and using the exploration-based matching method in the matching process. The effective use of the information transmission method to decompose the pattern graph is vital. A good decomposition method will reduce the amount of information to be transmitted, improving query efficiency. Simultaneously, introducing efficient algorithms to improve the accuracy of the results is critical.
(3) Simulation-based matching technique
In [23], the IncSimMatch algorithm and IncBMatch algorithm were compared against their corresponding batch algorithms, Match\(_s\) and Match\(_{bs}\), using real YouTube and citation network datasets. Figure 20a shows that the incremental algorithm performs better than its batch counterpart when the number of added or deleted edges does not exceed 40% of the total number of edges in the data graph. For the IncBMatch algorithm, Fig. 20b shows that the incremental algorithm is superior to the batch algorithm when the number of added or deleted edges does not exceed 20% of the total number of edges in the data graph.
Wickramaarachchi et al. [76] compared the proposed pruning algorithm, D-IDS, with the IncBMatch algorithm. IncBMatch pruned the data graph using restricted simulation, providing results that were less consistent with the structure of the pattern graph. Meanwhile, the D-IDS algorithm produced results that were more consistent with the pattern graph through binary simulation matching. The experimental results showed that the D-IDS algorithm could reduce the size of the data graph by an average of 60%. For data graphs with smaller diameters, the performance improvement was greater. The efficiency was also improved for data graphs with larger diameters, but not significantly. Kao et al. [41] compared the distributed incremental algorithm with the batch processing algorithm. The experimental results show that after the \(12^{th}\) update, the matching rate of the incremental algorithm was 3\(\sim \)10 times that of the batch processing algorithm, and more than 60% of the invalid updates could be filtered out.
In summary, the research on continuous subgraph matching based on the incremental technique is becoming increasingly extensive. Generally, the performance of the algorithm is considered from three perspectives. The first is to design an incremental algorithm that makes full use of the previous matching results to filter out invalid updates. The second is to design an efficient incremental subgraph matching algorithm to find candidate sets that may be matching results around the newly added edges. The third is to design a pruning strategy to further filter out invalid results in the matching candidate set.
Different application uses will require different approximation methods. For accuracy, the join-based method can obtain exact-matching results that are suitable for applications with strict requirements regarding the topology of the graph. In contrast, the join-based matching technique produces numerous intermediate results, resulting in lower efficiency. The exploration-based method will improve the matching efficiency, but accuracy will be lower; therefore, a matching method combining the two previous methods can be used to optimize performance. Further, graph simulation can be used for applications that do not have strict requirements for topology structure. Some extended graph simulation matching methods can obtain matching results with a higher correlation to the pattern graph, thereby compensating for the deficiencies of the join-based method and exploration-based method.
(4) Comprehensive comparison of incremental algorithms
Kim et al. [43] evaluated the performance of a data-centric representation (TurboFlux) against the query-centric representation (SJ-tree) and direct computation method (Graphflow), using the LSBench dataset, which was used in [17]. The experiment used both tree and graph queries. In the comparison of experimental results, both average elapsed time (\(M(\Delta g, q)\)) and storage cost were evaluated.
Figure 21 shows the performance results for the tree queries in LSBench. Notably, matching with an auxiliary data structure outperforms the direct computation method. The smaller query sizes noted better performance based on the average elapsed time. Further, data-centric presentation outperforms query-centric presentation. For an average elapsed time, TurboFlux outperformed SJ-tree by 77.30 \(\backsim \) 379.22 times and Graphflow by 515.01 \(\backsim \) 1,275.68 times regardless of query size. This is because SJ-tree generated a significant amount of partial solutions using the join operation. Since Graphflow used the direct computation method, it did not generate any intermediate results; therefore, TurboFlux was only compared with SJ-tree in terms of the storage cost. SJ-tree showed a notable storage cost problem. The average storage cost of SJ-tree was obviously larger than that of TurboFlux, up to 142.34 times. Evidently, TurboFlux could efficiently reduce the search space using DCG.
SJ-tree has a significant intermediate result size problem. Among them, the average size of the intermediate results of the sj tree is significantly larger than TurboFlux, reaching 142.34 times. Obviously, TurboFlux can effectively use DCG to reduce the search space.
From the above experimental results, it can be seen that maintaining intermediate results could increase the performance. Particularly, the data-centric representation has the best performance. TurboFlux consistently and significantly outperformed state-of-the-art methods for varying query sets, datasets, and insertion rates. The auxiliary data structures based on a graph can help us track affected vertices more quickly and obtain them more easily. In future research, new auxiliary data structures with higher performance and lower memory consumption should be considered.
Recently, Sun et al. [68] conducted an in-depth research on continuous subgraph matching. Instead of comparing tree and graph queries in [43], they also divided graph queries into sparse graph queries and dense graph queries. They found the latest algorithms do not consistently outperform the old ones and the direct computation incremental method runs faster than the incremental method with an auxiliary data structure in some cases. Specifically, (1) on tree queries, TurboFlux and SJ-Tree run much faster than Graphflow; (2) on sparse graph queries, Graphflow runs faster than TurboFlux; (3) on dense graph queries, Graphflow generally outperforms TurboFlux; (4) if edges are updated at the sparse regions in the data graph, Graphflow runs faster than TurboFlux.
5.2 Content-based change
Continuous subgraph matching algorithms based on content changes are generally compared in terms of query time and scalability. In [88], Gradin was compared with VF2, UpdAll, NaiveGrid, and UpdNo algorithms; VF2 is an indexless subgraph matching algorithm, UpdAll deploys query sharing into a multidimensional search tree, NaiveGrid is a grid index using the traditional verification algorithms, while UpdNo uses an inverted index. The experiment used the real dataset BCUBE, comprising the network architecture of a data center, where 3000 nodes were selected as the experimental dataset. As shown in Fig. 22, the experimental results demonstrated that the pruning rate and query time of the Gradin algorithm was similar to the UpdAll algorithm; however, the index construction time was 4 to 10 times that of the UpdAll algorithm. Simultaneously, the pruning rate of the Gradin algorithm is 10 times that of the UpdNo algorithm and 5 times that of the NaiveGrid algorithm. The Gradin algorithm uses join-based matching to speed up the graph matching process, giving it the fastest search speed in this case.
Continuous subgraph matching for content change, mainly used in the data centers, is not as widely researched as structural change. Generally, it is considered from the following two aspects. First, for applications with infrequent attribute updates, a join-based matching technique can be used, which is similar to static graph matching. Second, for applications with frequently updated attributes, an exploration-based matching method can be adopted, as a join-based matching method will produce numerous intermediate results.
6 Application analysis of continuous subgraph matching technique
In recent years, graph data have been widely used to describe complex and constantly changing relationships between various entities in the real world. Therefore, compared with the traditional static graph subgraph matching technique, the application scenarios of the continuous subgraph matching technique are more extensive. This section summarizes the practical applications of continuous subgraph matching techniques.
Potential drug trafficking in social network (Take from [23])
6.1 Criminal behavior analysis
Currently, criminal behaviors such as drug trafficking and terrorist attacks are causing serious harm worldwide. By establishing a behavioral relationship graph as a data graph, with people as nodes and activity relationships as edges, a criminal group behavioral relationship graph can be used to define a pattern graph. Using this pattern graph and a subgraph matching technique on a dynamic data graph, potentially criminal behavior can be discovered and predicted.
For example, a limited simulation matching technique was used on dynamic graphs to analyze potential drug trafficking groups in [23]. As shown in Fig. 23, where \(P_0\) is a pattern graph, with nodes representing criminals and edges representing the trafficking between them. The value of the edge label on the edge (AM, FW) is 3, indicating that the trafficking constraint needs to be satisfied from AM to FW within 3 hops. Based on the pattern graph, potential drug trafficking groups can be found by searching and matching in a large-scale dynamic behavior relationship graph, providing a strong basis for the detection of criminal behavior. It is reported that Palantir, a famous American intelligence company, also uses this technique to conduct investigations and analyze specific scenarios.
6.2 Road network monitoring
Urban road traffic monitoring is an important application of continuous subgraph matching. The urban road network can be expressed as a graph, where intersections are represented by nodes and road segments are represented by edges. Users can define pattern graphs based on typical road conditions that occur after traffic accidents and match the pattern graph with the dynamic road network data graph to achieve real-time monitoring of traffic accidents.
Taking the road network monitoring in [66] as an example, Fig. 24a represents typical road conditions that occur after a traffic accident, and the labels of the edges indicate the condition of the traffic (congested or smooth). In the matching process, the partial order relationship of time needs to be satisfied, which can be defined as the time relationship graph shown in Fig. 24b, where each node corresponds to an edge in Fig. 24a. Only when the subgraphs of the pattern graph and data graph match the order in the time domain, as defined in Fig. 24b, will they be considered as a final matching result. Each road traffic report can be regarded as an update to the road network graph data, and the updated part needs to be validated within the time window shown in Fig. 24c. Figure 24d, e represents two snapshots of the road network data graph at different points in time, where the numbers in parentheses represent time. The real-time traffic accident monitoring can be completed by subgraph matching between the event pattern graph (Fig. 24a) and the road network data graph that changes dynamically at different times (Fig. 24d, e). In this example, a subgraph in Fig. 24e matches the pattern graph.
6.3 Network security monitoring
With the rapid development of the Internet, network security issues have become more serious. Taking the network attack monitoring in [17] as an example, Fig. 25 shows three user-defined network attack behavior patterns. In the figure, nodes represent hosts and edges represent interaction relationships, such as communication between hosts and users during login. Using continuous subgraph matching between attack patterns and streaming graphs, both real and potential network attack events can be detected to predict network attack behavior.
6.4 Computational biology data analysis
In biology, molecular structures can be expressed as graphs, providing an important basis for studying the structure and function of biological tissues. In a protein interaction network, proteins react with certain enzymes causing mutations. By matching a known protein network pattern to a dynamic protein interaction network, the mutated protein structure can be found quickly. For example, Bader et al. [12] found that proteins with large betweenness and small connectivity are redundant proteins in human genes by calculating the connectivity and centrality of the protein interaction network and matching structures with known properties in a protein interaction network, which contained 18,000 proteins (nodes) and 44,000 protein interaction (edges). This undoubtedly provided an important basis for analyzing the function of genes and proteins.
7 Future work
As a widely used data model, dynamic graphs have theoretical significance and wide application prospects; therefore, the research of dynamic graph data matching has attracted increasing attention from academic and industrial circles. With the advent of big data, the scale of data has increased, data updates have become more frequent, and the relationships between data have become more complicated. This undoubtedly has brought new challenges and opportunities to the research and application of continuous subgraph matching. We propose a list of promising future directions as follows.
Distributed Parallel Graph Process Faced with the rapid increase in the scale of graph data, the scalability of matching techniques for large-scale graph data needs to be improved. Therefore, methods to realize parallel distributed processing of graph matching, which are based on mainstream distributed parallel graph processing framework systems, such as Pregel and Giraph, have to be studied. These mainstream distributed parallel graph processing framework systems usually adopt a vertex-centric processing mode. Each node, as a basic computing unit in the framework, stores and processes a partition of the graph data, while in a block-centric model, the basic calculation unit is a block, which stores and processes a connected subgraph of the data graph. The two distributed parallel graph processing models have their own advantages and disadvantages, and researchers can apply the most suited model according to their application needs.
Incremental Algorithm For frequently updated graph data, the snapshot-based matching method is not ideal for meeting the real-time requirements of continuous subgraph matching. Therefore, efficient matching methods and query optimization strategies based on the feature of dynamic graphs have to be studied. More and more incremental subgraph matching algorithms have been proposed which use different auxiliary data structures [43], or filtering strategies [54] or matching order optimization strategies [67]. In order to better perform the continuous subgraph matching, our main focus is still on the construction of the incremental algorithm with some optimization strategies to improve matching efficiency.
Matching Technique Join-based matching technique is an effective method to achieve continuous subgraph matching. However, extracting high-quality features from the pattern graph or data graph is one of the core problems faced by this technique. On the one hand, to avoid failure from data graph updates, current studies generally choose to use simple subgraphs, such as single-edge subgraphs or double-edge subgraphs, as the characteristics of the data graph. However, such simple subgraphs do not have strong identifiable characteristics, leading to many intermediate matching results and an increased processing cost for the subgraph join phase. Therefore, methods to extract more recognizable features and design efficient feature maintenance schemes should be the focus of future research. On the other hand, the exploration-based continuous subgraph matching method has its advantages but delivers low accuracy matching results. To solve this problem, further research can be conducted to combine exploration-based and join-based continuous subgraph matching techniques. The studies should focus on making optimal use of the advantages of the respective techniques to compensate for the deficiencies. Several studies have focused on the simulation matching technique that avoids complex subgraph isomorphic matching calculations. Further research on this topic would be advantageous. In addition, designing a graph matching similarity measurement model based on specific requirements and developing a more efficient graph matching approximation algorithm are future research considerations.
Pattern Evolving Most of the current research on continuous subgraph matching is directed toward situations where the pattern graph remains unchanged and the data graph changes with time. However, in real life, changes to the pattern graph are also very common. For example, in network security, viruses are often mutated, and network attack patterns are constantly evolving; in computational biology, protein denaturation and virus mutations also occur occasionally. [86] studied the incremental algorithm of graph pattern matching when the data graph was unchanged, but the pattern graph changed dynamically. And Zhang et al. [84] modeled network traffic as a large graph and attack behaviors as evolving pattern graphs. And then the abnormal attacks can be detected through continuous subgraph matching. Therefore, researchers can explore the incremental processing technique required in these areas; furthermore, techniques that accommodate the changes in both pattern graphs and data graphs can be explored to expand the application field of continuous subgraph matching techniques.
Multi-query Answer Most existing continuous subgraph matching studies process only one query at a time. However, owing to large-scale and constantly evolving graphs, it is more practical to use multiple queries to monitor and detect continuous patterns of interest. Pugliese et al. [59] utilized a merged view of multiple query graphs to update results incrementally. Zervakis et al. [83] designed a query graph clustering algorithm to handle a large number of continuous queries. Mhedhbi et al. [52] proposed a general greedy optimizer to share computation among multiple instances of continuous queries. Although some state-of-the-art methods exist, how to reduce space consumption and time consumption, and how to quickly detect the queries affected by updates are still the main problems. All these methods can be further improved via a better filtering strategy and matching order or more efficient auxiliary data structures.
8 Conclusion
In this paper, we extensively investigate the topic of continuous subgraph matching over dynamic graphs in 20 research articles published between 2009 and 2020. We first classify these articles according to different problem indicators. Then, we review and discuss two different types of continuous subgraph matching approaches based on the different update methods of the dynamic graph. In addition, the performance of each algorithm is discussed. Finally, we point out the future research directions and main challenges. In summary, our survey outlines the start-of-the-art research achievements about continuous subgraph matching, which will give researchers a thorough understanding.
References
Apache girph[db/ol]. http://incubator.apache.org/giraph
Facebook quarterly update. http://bit.ly/2bim30d
Neo4j. https://neo4j.com/
Orientdb. http://orientdb.com/orientdb/
Postech database lab. nasa, yeast, and human datasets.[2012-08-27].[2014-11-20]. http://dtp.nci.nih.gov/docs/aids/aids_data.html
Reasoning on rdf streams. [online]: http://streamreasoning.org/publication (2013)
Verizon. http://www.verizonenterprise.com/resources/reports.pdf (2016)
Reality mining dataset[db/ol]. http://reality.media.mit.edu (2017)
Aggarwal CC, Wang H. (eds.) (2010) Managing and Mining Graph Data, Advances in Database Systems, vol. 40. Springer. https://doi.org/10.1007/978-1-4419-6045-0
Ammar K, McSherry F, Salihoglu S, Joglekar M (2018) Distributed evaluation of subgraph queries using worst-case optimal and low-memory dataflows. Proc VLDB Endow 11(6):691–704
Aridhi S, Montresor A, Velegrakis Y (2017) BLADYG: a graph processing framework for large dynamic graphs. Big Data Res 9:9–17. https://doi.org/10.1016/j.bdr.2017.05.003
Bader DA, Madduri K (2007) A graph-theoretic analysis of the human protein-interaction network using multicore parallel algorithms. In: Proceedings of 21th international parallel and distributed processing symposium, pp 1–8. https://doi.org/10.1109/IPDPS.2007.370445
Barbieri DF, Braga D, Ceri S, Valle ED, Grossniklaus M (2010) C-SPARQL: a continuous query language for RDF data streams. Int J Semant Comput 4(1):3–25. https://doi.org/10.1142/S1793351X10000936
Boshmaf Y, Muslukhov I, Beznosov K, Ripeanu M (2011) The socialbot network: when bots socialize for fame and money. In: Proceedings of twenty-seventh annual computer security applications conference, pp 93–102. https://doi.org/10.1145/2076732.2076746
Cheng J, Ke Y, Ng W, Lu A (2007) Fg-index: towards verification-free query processing on graph databases. In: Proceedings of international conference on management of data, pp 857–872. https://doi.org/10.1145/1247480.1247574
Choudhury S, Holder LB, Feo J, Jr, GC (2013) Fast search for dynamic multi-relational graphs. In: Proceedings of the workshop on dynamic networks management and mining, pp 1–8. https://doi.org/10.1145/2489247.2489251
Choudhury S, Holder LB, Jr, GC, Agarwal K, Feo J (2015) A selectivity based approach to continuous pattern detection in streaming graphs. In: Proceedings of the 18th international conference on extending database technology, pp 157–168. https://doi.org/10.5441/002/edbt.2015.15
Choudhury S, Holder LB, Jr, GC, Mackey P, Agarwal K, Feo J (2014) Query optimization for dynamic graphs. CoRR abs/1407.3745. http://arxiv.org/abs/1407.3745
Consortium TU (2017) Uniprot: the universal protein knowledgebase. Nucleic Acids Res. 45(Database-Issue), D158–D169. https://doi.org/10.1093/nar/gkw1099
Cordella LP, Foggia P, Sansone C, Vento M (2004) A (sub)graph isomorphism algorithm for matching large graphs. IEEE Trans Pattern Anal Mach Intell 26(10):1367–1372. https://doi.org/10.1109/TPAMI.2004.75
Fan W, Fan Z, Tian C, Dong XL (2015) Keys for graphs. In: Proceedings of VLDB Endowment 8(12), 1590–1601. http://www.vldb.org/pvldb/vol8/p1590-fan.pdf
Fan W, Hu C, Tian C (2017) Incremental graph computations: doable and undoable. In: Proceedings of the 2017 international conference on management of data, pp. 155–169. https://doi.org/10.1145/3035918.3035944
Fan W, Li J, Luo J, Tan Z, Wang X, Wu Y (2011) Incremental graph pattern matching. In: Proceedings of the 2011 international conference on management of data, pp. 925–936. https://doi.org/10.1145/1989323.1989420
Fan W, Wang X, Wu Y, Xu J (2015) Association rules with graph patterns. Proc VLDB Endow 8(12):1502–1513
Fan W, Wu Y, Xu J (2016) Functional dependencies for graphs. In: Proceedings of the 2016 international conference on management of data, pp. 1843–1857. https://doi.org/10.1145/2882903.2915232
Fan W, Xu J, Wu Y, Yu W, Jiang J, Zheng Z, Zhang B, Cao Y, Tian C (2017) Parallelizing sequential graph computations. In: Proceedings of the 2017 international conference on management of data, pp. 495–510. https://doi.org/10.1145/3035918.3035942
Fang Y, Huang X, Qin L, Zhang Y, Zhang W, Cheng R, Lin X (2020) A survey of community search over big graphs. VLDB J 29(1):353–392. https://doi.org/10.1007/s00778-019-00556-x
Fournier-Viger P, He G, Cheng C, Li J, Zhou M, Lin JC, Yun U (2020) A survey of pattern mining in dynamic graphs. WIREs Data Mining Knowl Discov. https://doi.org/10.1002/widm.1372
Gao J, Zhou C, Yu JX (2016) Toward continuous pattern detection over evolving large graph with snapshot isolation. VLDB J 25(2):269–290. https://doi.org/10.1007/s00778-015-0416-z
Gao J, Zhou C, Zhou J, Yu JX (2014) Continuous pattern detection over billion-edge graph using distributed framework. In: Proceedings of the 30th international conference on data engineering, pp 556–567. https://doi.org/10.1109/ICDE.2014.6816681
Giugno R, Shasha DE (2002) Graphgrep: a fast and universal method for querying graphs. In: Proceedings of the 16th international conference on pattern recognition, pp 112–115. https://doi.org/10.1109/ICPR.2002.1048250
Gong NZ, Xu W, Huang L, Mittal P, Stefanov E, Sekar V, Song D (2012) Evolution of social-attribute networks: measurements, modeling, and implications using google+. In: Proceedings of the 12th internet measurement conference, pp 131–144. https://doi.org/10.1145/2398776.2398792
Hajlaoui JE, Omri MN, Benslimane D (2017) Performance and scalability appraisal of four directed weighted graph matching algorithms: A survey. In: Proceedings of the 14th international conference on computer systems and applications, pp 392–398. https://doi.org/10.1109/AICCSA.2017.50
Han W, Lee J, Lee J (2013) Turbo\({}_{\text{iso}}\): towards ultrafast and robust subgraph isomorphism search in large graph databases. In: Proceedings of the 2013 international conference on management of data, pp 337–348. https://doi.org/10.1145/2463676.2465300
He H, Singh AK (2006) Closure-tree: An index structure for graph queries. In: Proceedings of the 22nd international conference on data engineering, p 38. https://doi.org/10.1109/ICDE.2006.37
He H, Singh AK (2008) Graphs-at-a-time: query language and access methods for graph databases. In: Proceedings of the 2008 international conference on management of data, pp 405–418. https://doi.org/10.1145/1376616.1376660
Jiang H, Wang H, Yu PS, Zhou S (2007) Gstring: a novel approach for efficient search in graph databases. In: Proceedings of the 23rd international conference on data engineering, pp 566–575. https://doi.org/10.1109/ICDE.2007.367902
Jiang N, Jin Y, Skudlark A, Hsu W, Jacobson G, Prakasam S, Zhang Z (2012) Isolating and analyzing fraud activities in a large cellular network via voice call graph analysis. In: Proceedings of the 10th international conference on mobile systems, pp 253–266. https://doi.org/10.1145/2307636.2307660
Jing Y, Yanbing L, Yu Z, Mengya L, Jianlong T, Li G (2015) Survey on large-scale graph pattern matching. J Comput Res Dev 52(2):391–409
Kankanamge C, Sahu S, Mhedbhi A, Chen J, Salihoglu S (2017) Graphflow: an active graph database. In: Proceedings of the 2017 international conference on management of data, pp 1695–1698. https://doi.org/10.1145/3035918.3056445
Kao J, Chou J (2016) Distributed incremental pattern matching on streaming graphs. In: Proceedings of the workshop on high performance graph processing, pp 43–50. https://doi.org/10.1145/2915516.2915519
Khurana U, Deshpande A (2013) Efficient snapshot retrieval over historical graph data. In: Proceedings of the 29th international conference on data engineering, pp 997–1008. https://doi.org/10.1109/ICDE.2013.6544892
Kim K, Seo I, Han W, Lee J, Hong S, Chafi H, Shin H, Jeong G (2018) TurboFlux: a fast continuous subgraph matching system for streaming graph data. In: Proceedings of the 2018 international conference on management of data, pp 411–426. https://doi.org/10.1145/3183713.3196917
Kou Y, Shen D, Snell Q, Li D, Nie T, Yu G, Ma S (2020) Efficient team formation in social networks based on constrained pattern graph. In: Proceedings of ICDE, Dallas, TX, USA, April 20–24, pp 889–900
Kuramochi M, Karypis G (2001) Frequent subgraph discovery. In: Proceedings of ICDM, 29 November–2 December, San Jose, California, USA, pp 313–320
Lai L, Qing Z, Yang Z et al (2019) Distributed subgraph matching on timely dataflow. Proc VLDB Endow 12(10):1099–1112
Lee J, Han W, Kasperovics R, Lee J (2012) An in-depth comparison of subgraph isomorphism algorithms in graph databases. Proc VLDB Endow 6(2):133–144
Li Y, Zou L, Özsu MT, Zhao D (2019) Time constrained continuous subgraph search over streaming graphs. In: Proceedings of the 35th international conference on data engineering, pp 1082–1093
Low Y, Gonzalez J, Kyrola A, Bickson D, Guestrin C, Hellerstein JM (2010) Graphlab: a new framework for parallel machine learning. In: Proceedings of the twenty-sixth conference on uncertainty in artificial intelligence, pp 340–349. https://dslpitt.org/uai/displayArticleDetails.jsp?mmnu=1 &smnu=2 &article_id=2126
Ma S, Cao Y, Fan W, Huai J, Wo T (2011) Capturing topology in graph pattern matching. Proc VLDB Endow 5(4):310–321
Malewicz G, Austern MH, Bik AJC, Dehnert JC, Horn I, Leiser N, Czajkowski G (2010) Pregel: a system for large-scale graph processing. In: Proceedings of the international conference on management of data, pp 135–146. https://doi.org/10.1145/1807167.1807184
Mhedhbi A, Kankanamge C, Salihoglu S (2021) Optimizing one-time and continuous subgraph queries using worst-case optimal joins. ACM Trans Database Syst. 46(2), 6:1–6:45. https://doi.org/10.1145/3446980
Milner R (1989) Communication and concurrency. PHI Series in computer science. Prentice Hall
Min S, Park SG, Park K, Giammarresi D, Italiano GF, Han W (2021) Symmetric continuous subgraph matching with bidirectional dynamic programming. Proc VLDB Endow 14(8):1298–1310
Mondal J, Deshpande A (2012) Managing large dynamic graphs efficiently. In: Proceedings of the 2012 international conference on management of data, pp 145–156. https://doi.org/10.1145/2213836.2213854
Mondal J, Deshpande A (2016) CASQD: continuous detection of activity-based subgraph pattern queries on dynamic graphs. In: Proceedings of the 10th international conference on distributed and event-based systems, pp 226–237. https://doi.org/10.1145/2933267.2933316
Ngo HQ, Porat E, Ré C, Rudra A (2018) Worst-case optimal join algorithms. J ACM 65(3), 16:1–16:40. https://doi.org/10.1145/3180143
Ngo HQ, Ré C, Rudra A (2013) Skew strikes back: new developments in the theory of join algorithms. SIGMOD Rec 42(4):5–16. https://doi.org/10.1145/2590989.2590991
Pugliese A, Bröcheler M, Subrahmanian VS, Ovelgönne M (2014) Efficient multiview maintenance under insertion in huge social networks. ACM Trans Web 8(2), 10:1–10:32. https://doi.org/10.1145/2541290
Raghavendra R, Lobo J, Lee K (2012) Dynamic graph query primitives for SDN-based cloudnetwork management. In: Proceedings of the the first workshop on Hot topics in software defined networks, pp 97–102. https://doi.org/10.1145/2342441.2342461
Ren X, Wang J (2015) Exploiting vertex relationships in speeding up subgraph isomorphism over large graphs. Proc VLDB Endow 8(5), 617–628. http://www.vldb.org/pvldb/vol8/p617-ren.pdf
Ren X, Wang J (2016) Multi-query optimization for subgraph isomorphism search. Proc VLDB Endow 10(3), 121–132. http://www.vldb.org/pvldb/vol10/p121-ren.pdf
Shang H, Zhang Y, Lin X, Yu JX (2008) Taming verification hardness: an efficient algorithm for testing subgraph isomorphism. Proc VLDB Endow 1(1), 364–375. http://www.vldb.org/pvldb/vol1/1453899.pdf
Shao B, Wang H, Li Y (2013) Trinity: a distributed graph engine on a memory cloud. In: Proceedings of the international conference on management of data, pp 505–516. https://doi.org/10.1145/2463676.2467799
Shasha DE, Wang JT, Giugno R (2002) Algorithmics and applications of tree and graph searching. In: Proceedings of the twenty-first symposium on principles of database systems, pp 39–52. https://doi.org/10.1145/543613.543620
Song C, Ge T, Chen CX, Wang J (2014) Event pattern matching over graph streams. Proc VLDB Endow 8(4):413–424
Sun S, Luo Q (2022) Subgraph matching with effective matching order and indexing. IEEE Trans Knowl Data Eng 34(1):491–505. https://doi.org/10.1109/TKDE.2020.2980257
Sun X, Sun S, Luo Q, He B (2022) An in-depth study of continuous subgraph matching (complete version). CoRR abs/2203.06913. https://doi.org/10.48550/arXiv.2203.06913
Sun Z, Wang H, Wang H, Shao B, Li J (2012) Efficient subgraph matching on billion node graphs. Proc VLDB Endow 5(9):788–799
Tian Y, Patel JM (2008) TALE: a tool for approximate large graph matching. In: Proceedings of the 24th international conference on data engineering, pp 963–972. https://doi.org/10.1109/ICDE.2008.4497505
Ugander J, Karrer B, Backstrom L, Marlow C (2011) The anatomy of the Facebook social graph. CoRR abs/1111.4503. http://arxiv.org/abs/1111.4503
Valiant LG (1990) A bridging model for parallel computation. Commun ACM 33(8):103–111. https://doi.org/10.1145/79173.79181
Veldhuizen TL (2014) Triejoin: a simple, worst-case optimal join algorithm. In: Proceedings of the 17th international conference on database theory, pp 96–106. https://doi.org/10.5441/002/icdt.2014.13
Wang AH (2010) Don’t follow me - spam detection in twitter. In: Proceedings of the international conference on security and cryptography, pp 142–151
Wang C, Chen L (2009) Continuous subgraph pattern search over graph streams. In: Proceedings of the 25th international conference on data engineering, pp 393–404. https://doi.org/10.1109/ICDE.2009.132
Wickramaarachchi C, Kannan R, Chelmis C, Prasanna VK (2016) Distributed exact subgraph matching in small diameter dynamic graphs. In: Proceedings of the 2016 international conference on big data, pp 3360–3369. https://doi.org/10.1109/BigData.2016.7840996
Xenarios I, Salwínski L, Duan XJ, Higney P, Kim S, Eisenberg DS (2002) Dip, the database of interacting proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res 30(1):303–305. https://doi.org/10.1093/nar/30.1.303
Yan D, Cheng J, Lu Y, Ng W (2014) Blogel: a block-centric framework for distributed computation on real-world graphs. Proc VLDB Endow 7(14):1981–1992
Yan X, Han J (2002) gspan: graph-based substructure pattern mining. In: Proceedings of the 2002 international conference on data mining, pp 721–724. https://doi.org/10.1109/ICDM.2002.1184038
Yan X, Yu PS, Han J (2004) Graph indexing: a frequent structure-based approach. In: Proceedings of the 2004 international conference on management of data, pp 335–346. https://doi.org/10.1145/1007568.1007607
Yang J, Jin W (2011) Br-index: an indexing structure for subgraph matching in very large dynamic graphs. In: Proceedings of the 23rd international conference on scientific and statistical database management, vol. 6809, pp 322–331. https://doi.org/10.1007/978-3-642-22351-8_20
Yang J, Zhang S, Jin W (2011) DELTA: indexing and querying multi-labeled graphs. In: Proceedings of the 20th conference on information and knowledge management, pp 1765–1774. https://doi.org/10.1145/2063576.2063832
Zervakis L, Setty V, Tryfonopoulos C, Hose K (2020) Efficient continuous multi-query processing over graph streams. In: Proceedings of the 23rd international conference on extending database technology, pp 13–24. https://doi.org/10.5441/002/edbt.2020.03
Zhang Q, Guo D, Zhao X, Guo A (2019) On continuously matching of evolving graph patterns. In: Proceedings of the 28th international conference on information and knowledge management, pp 2237–2240. https://doi.org/10.1145/3357384.3358101
Zhang S, Li S, Yang J (2009) GADDI: distance index based subgraph matching in biological networks. In: Proceedings of the 12th international conference on extending database technology, vol. 360, pp 192–203. https://doi.org/10.1145/1516360.1516384
Zhang LX Wang WP, GJWJ (2015) Pattern graph change oriented incremental graph pattern matching. J Softw 26(11)
Zhao P, Han J (2010) On graph query optimization in large networks. Proc VLDB Endow 3(1):340–351
Zong B, Raghavendra R, Srivatsa M, Yan X, Singh AK, Lee K (2014) Cloud service placement via subgraph matching. In: Proceedings of the 30th international conference on data engineering, pp 832–843. https://doi.org/10.1109/ICDE.2014.6816704
Acknowledgements
This work is partially supported by National Key Research and Development Program No. 2018YFE0207600, National Natural Science Foundation of China under Grant No. U19B2024, National Natural Science Foundation of China under Grant No. 61872446, The Science and Technology Innovation Program of Hunan Province under grant No. 2020RC4046, and Postgraduate Scientific Research Innovation Project of Hunan Province under Grant No.CX20210038.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, X., Zhang, Q., Guo, D. et al. A survey of continuous subgraph matching for dynamic graphs. Knowl Inf Syst 65, 945–989 (2023). https://doi.org/10.1007/s10115-022-01753-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10115-022-01753-x