Abstract
Nattu kozhi (Red jungle fowl—Gallus gallus domesticus) is a breed of chickens that is native to the southern regions of India, Vellore. These chickens are known for their high-quality feathers, but when erroneously disposed, it transmits significant disease threat to human as well as animal health, such as fowl cholera, allergies, salmonellosis, histoplasmosis and pollutes the environment. The identification of amino acids from keratin, a fibrous protein found in feathers, is an important step in the development of sustainable and renewable sources of animal-derived protein. In this study, we aimed to identify various amino acids from the Nattu kozhi feather keratin hydrolysates. The keratin degrading bacteria was first isolated using serial dilution and baiting technique, wherein feather was placed on Nutrient agar and Luria Bertani agar plates and incubated at 37 °C for 24–48 h. The bacteria which degraded the feather from Luria bertani agar plate was then subjected to further characterization analysis. Additionally, the isolated colonies were incubated with Nattu kozhi feathers in nutrient and Luria Bertani broth for 20–21 days to observe degradation of feathers. Once the degradation was successful, a microscopic analysis aids in identifying the Bacillus bacterial species that assisted in the degradation process. Various biochemical tests were also performed for the same. The feather hydrolysate sample was passed through various chromatographic techniques like paper and TLC in order to identify the amino acids. Various amino acids, such as cysteine, glycine, methionine and lysine, were quantified using high-performance liquid chromatography. FT-IR and X-ray diffraction studies gave an insights into the functional group and nanoparticle conformation of the protein and amino acid. Molecular docking studies resulted that, the ligand [3-(trifluoromethyl) phenyl urea] has − 5.2 kcal/mol affinity towards keratin and may aid in catalysing the degradation reaction. This eco-friendly, sustainable technology transforms and mitigates pollution and toxicity from feather waste, enabling the recovery of amino acids with applications and commercialization potential across various industrial sectors.
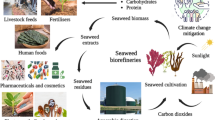
Graphical abstract





























Similar content being viewed by others
Abbreviations
- 2D:
-
Two dimension
- 3D:
-
Three dimension
- AMC:
-
Acetylmethylcarbinol
- Asp:
-
Aspartic acid
- CID:
-
Compound identifier
- EDTA:
-
Ethylene diamine tetra-acetic acid
- FT-IR:
-
Fourier transform infrared spectroscopy
- Glu:
-
Glutamic acid
- GOR:
-
Garnier–Osguthorpe–Robson
- HEPES:
-
N-(2-hydroxyethyl)piperazine-Nʹ-2-ethanesulfonic acid
- HPLC:
-
High performance liquid chromatography
- IMVIC:
-
Indole/methyl red/Voges Prousker/citrate utilization
- KH2PO4:
-
Potassium dihydrogen phosphate
- LB:
-
Luria Bertani broth
- NA:
-
Nutrient agar
- NaOH:
-
Sodium hydroxide
- NB:
-
Nutrient broth
- PDB:
-
Protein data bank
- Phe:
-
Phenylalanine
- RCSB:
-
Research collaboratory for structural bioinformatics
- Rf:
-
Retardation/retention factor
- RPM:
-
Revolution per minute
- SBD:
-
Substrate-binding domain
- TES:
-
N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid
- TLC:
-
Thin layer chromatography
- UV:
-
Ultra violet
- UV–Vis:
-
UV–visible spectroscopy
- Val:
-
Valine
- XRD:
-
X-ray diffraction
References
Ali A, Chiang YW, Santos RM (2022) X-ray diffraction techniques for mineral characterization: a review for engineers of the fundamentals, applications, and research directions. Minerals 12:205. https://doi.org/10.3390/min12020205
Alkhuder K (2022) Attenuated total reflection-Fourier transform infrared spectroscopy: a universal analytical technique with promising applications in forensic analyses. Int J Legal Med 136:1717–1736. https://doi.org/10.1007/s00414-022-02882-2
Bele AA, Khale A (2011) An overview on thin layer chromatography Archana A. Bele* and Anubha Khale H. K. College of Pharmacy, Jogeshwari (W), Mumbai, Maharashtra. India J Pharm Sci 2:256–267
Bhutia MO, Thapa N, Tamang JP (2021) Molecular characterization of bacteria, detection of enterotoxin genes, and screening of antibiotic susceptibility patterns in traditionally processed meat products of Sikkim, India. Front Microbiol. https://doi.org/10.3389/fmicb.2020.599606
Bischoff R, Schlüter H (2012) Amino acids: chemistry, functionality and selected non-enzymatic post-translational modifications. J Proteomics 75:2275–2296. https://doi.org/10.1016/j.jprot.2012.01.041
Boone C, Adamec J (2016) 10—Top–down proteomics. In: Ciborowski P, Silberring J (eds) Proteomic profiling and analytical chemistry, 2nd edn. Elsevier, Boston, pp 175–191
Cai L (2014) Thin layer chromatography. Curr Protoc Essent Lab Tech 8:6.3.1-6.3.18. https://doi.org/10.1002/9780470089941.et0603s08
Coskun O (2016) Separation techniques: chromatography. North Clin Istanb 3:156–160. https://doi.org/10.14744/nci.2016.32757
Donato RK, Mija A (2020) Keratin associations with synthetic, biosynthetic and natural polymers: an extensive review. Polymers 12:32. https://doi.org/10.3390/polym12010032
Eldirany SA, Ho M, Hinbest AJ et al (2019) Human keratin 1/10–1B tetramer structures reveal a knob-pocket mechanism in intermediate filament assembly. EMBO J 38:e100741. https://doi.org/10.15252/embj.2018100741
Forgács G, Alinezhad S, Mirabdollah A et al (2011) Biological treatment of chicken feather waste for improved biogas production. J Environ Sci China 23:1747–1753. https://doi.org/10.1016/s1001-0742(10)60648-1
Fraser RDB, Parry DAD (2009) The role of β-sheets in the structure and assembly of keratins. Biophys Rev 1:27. https://doi.org/10.1007/s12551-008-0005-0
Guentzel MN (1996) Escherichia, Klebsiella, Enterobacter, Serratia, Citrobacter, and Proteus. In: Baron S (ed) Medical microbiology. 4th ed. Chapter 26. University of Texas medical Branch at Galveston, Galveston
Kaur M, Bhari R, Singh RS (2021) Chicken feather waste-derived protein hydrolysate as a potential biostimulant for cultivation of mung beans. Biologia (bratisl) 76:1807–1815. https://doi.org/10.1007/s11756-021-00724-x
Kouza M, Faraggi E, Kolinski A, Kloczkowski A (2017) The GOR method of protein secondary structure prediction and its application as a protein aggregation prediction tool. In: Zhou Y, Kloczkowski A, Faraggi E, Yang Y (eds) Prediction of protein secondary structure. Springer, New York, pp 7–24
Li Q (2019) Progress in microbial degradation of feather waste. Front Microbiol. https://doi.org/10.3389/fmicb.2019.02717
Ljutov V (1959) Technique of indole test. Acta Pathol Microbiol Scand 46:349–360. https://doi.org/10.1111/j.1699-0463.1959.tb01106.x
Ljutov V (1961) Technique of methyl red test. Acta Pathol Microbiol Scand 51:369–380. https://doi.org/10.1111/j.1699-0463.1961.tb00375.x
Matsuda R, Rodriguez E, Suresh D, Hage DS (2015) Chromatographic immunoassays: strategies and recent developments in the analysis of drugs and biological agents. Bioanalysis 7:2947–2966. https://doi.org/10.4155/bio.15.206
Meng X-Y, Zhang H-X, Mezei M, Cui M (2011) Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des 7:146–157. https://doi.org/10.2174/157340911795677602
Mozhiarasi V, Natarajan TS (2022) Slaughterhouse and poultry wastes: management practices, feedstocks for renewable energy production, and recovery of value added products. Biomass Convers Biorefinery. https://doi.org/10.1007/s13399-022-02352-0
Neda I, Vlazan P, Pop RO et al (2012) Peptide and amino acids separation and identification from natural products. In: Krull IS (ed) Analytical chemistry. IntechOpen, Rijeka
O’Toole GA (2016) Classic spotlight: how the gram stain works. J Bacteriol 198:3128. https://doi.org/10.1128/jb.00726-16
Qiu J, Wilkens C, Barrett K, Meyer AS (2020) Microbial enzymes catalyzing keratin degradation: classification, structure, function. Biotechnol Adv 44:107607. https://doi.org/10.1016/j.biotechadv.2020.107607
Sharma S, Gupta A (2016) Sustainable management of keratin waste biomass: applications and future perspectives. Braz Arch Biol Technol 59:e16150684. https://doi.org/10.1590/1678-4324-2016150684
Srivastava B, Khatri M, Singh G, Arya SK (2020) Microbial keratinases: an overview of biochemical characterization and its eco-friendly approach for industrial applications. J Clean Prod 252:119847. https://doi.org/10.1016/j.jclepro.2019.119847
Werkman CH (1930) An improved technic for the Voges–Proskauer test. J Bacteriol 20:121–125. https://doi.org/10.1128/jb.20.2.121-125.1930
Windt X, Scott EL, Seeger T et al (2022) Fourier transform infrared spectroscopy for assessing structural and enzymatic reactivity changes induced during feather hydrolysis. ACS Omega 7:39924–39930. https://doi.org/10.1021/acsomega.2c04216
Acknowledgements
We would like to acknowledge Hi-Media and Sigma-Aldrich chemicals for research support to our Science, Innovation and Society Lab. We would like to acknowledge the School of Advances Sciences from VIT University, Vellore for their valuable support in analyzing FTIR, XRD spectral and HPLC analysis. The authors express their gratitude, to Dr. G. Viswanathan, VIT University, Vellore, India for his constant motivation, encouragement and support.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
Professor Dr. Suneetha Vuppu made a pivotal role in developing the manuscript’s framework, provided multifaceted contributions extended to manuscript drafting, articulating the research findings and ensuring clarity, conducting microscopic analyses, biochemical characterization studies, leading FTIR and XRD characterization studies, conducting in silico docking studies, and crafting Biorender diagrams. The manuscript was read and analytically revised by all authors for important intellectual content. Sathvika Kamaraj contributed significantly to the manuscript by conceptualizing its structure, conducting microscopic analysis, performing biochemical characterizations, executing FTIR and XRD studies, utilizing chromatographic techniques such as TLC, performing in silico docking studies, and creating Biorender diagrams. Thushar Suresh helped in microscopic analysis, overseeing biochemical characterization studies, XRD characterization studies, employing chromatographic techniques like TLC, conducting in silico docking studies, and Biorender diagrams.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kamaraj, S., Suresh, T. & Vuppu, S. A sustainable approach for degrading Vellore Nattu kozhi feather waste, analytical amino acid profiling and its computational studies. Clean Techn Environ Policy (2024). https://doi.org/10.1007/s10098-024-02840-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10098-024-02840-z