Abstract
Purpose
Data on the molecular epidemiology of methicillin-resistant Staphylococcus aureus isolates from patients with bacteraemia in Slovenia are lacking. The aim of this study was to phenotypically and genotypically investigate 82 MRSA strains isolated from patients with bloodstream infections in central Slovenia between 2019 and 2022.
Methods
Whole-genome sequencing of selected strains was performed to characterize the strains based on sequence typing, antimicrobial resistance, toxin, and virulence factors genes.
Results
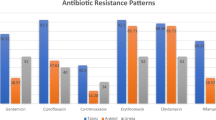
Most MRSA carried SCCmec II (63.4%), followed by SCCmec IV (34.1%) and SCCmec V (2.5%). A high proportion of strains belonging to the ST225 lineage (45.1%) was observed, followed by ST97 (18.3%), ST2883 (15.9%), ST22 (9.8%), ST5 (3.7%), and the ST1, ST398 and ST45 lineages (2.4% each). Sixteen different spa types were identified, predominantly ST225-t003 (31.7%), ST97-t359 (15.9%), and ST2883-t4336 (14.6%). None of the strains carried Panton-Valentine leukocidin, exfoliative toxins, or toxic shock toxin. All MRSA strains were susceptible to linezolid, rifampicin, vancomycin, teicoplanin, and trimethoprim-sulfamethoxazole. MRSA strains were resistant to erythromycin, clindamycin, tetracycline and gentamicin, with a frequency of 74.4%, 74.4%, 8.5%, and 1.2%, respectively.
Conclusion
This study demonstrates that bacteraemia in central Slovenia is caused by diverse MRSA lineages. Identification of newly emerged lineages should be followed in the future to detect changes in the molecular epidemiology of MRSA in our country.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The global epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) has changed due to different sources of MRSA strains, including hospital-associated MRSA (HA-MRSA), community-associated MRSA (CA-MRSA), and livestock-associated MRSA (LA-MRSA) worldwide. Epidemiologic distinctions between HA-MRSA, CA-MRSA, and LA-MRSA are blurred because MRSA clones circulate in hospital settings, communities, and livestock environment [1, 2].
MRSA is an important nosocomial pathogen that causes bloodstream infections (BSIs), which significantly increase morbidity and mortality, antibiotic use, and the cost of treatment [1, 3, 4]. In the United States, the USA300 clone is the most common strain causing nosocomial MRSA bacteraemia, whereas in Europe, most nosocomial MRSA infections continue to be caused by HA-MRSA genotypes (e.g. ST5, ST239, ST247, ST250, ST22, ST45), except in countries with industrial swine production, where LA-MRSA ST398 is predominant [1, 2, 5,6,7,8].
MRSA is well controlled in Slovenian hospitals, but unfortunately the molecular epidemiology of MRSA in Slovenia is poorly documented, even if the molecular epidemiology of MRSA in Europe has been extensively documented [6,7,8]. At the national level, we only have data on antimicrobial resistance and prevalence of MRSA isolates from the Slovenian National Antimicrobial Susceptibility Testing Committee (SKUOPZ) and from the European Resistance Surveillance Network (EARS-Net). According to the data from EARS-Net, the prevalence of invasive MRSA isolates from blood cultures or cerebrospinal fluid among all S. aureus isolates in Slovenia in 2021 was 7.8% [9]. According to latest available data from SKUOPZ the prevalence of MRSA isolates from clinical samples among all S. aureus isolates in Slovenia in 2017 was 7.7% [10].
In addition, limited data on virulence gene profiles and clonality of S. aureus and MRSA have been reported in Slovenia [11,12,13,14,15]. There are very few published reports investigating the molecular epidemiology of MRSA bloodstream infections in Slovenia. To our knowledge, the last publication on the genetic characterization of 10 invasive MRSA isolates from blood cultures is from 2006, and the predominant spa type was t041 [16, 17].
For these reasons, we performed whole-genome sequencing of MRSA isolates recovered from blood cultures between 2019 and 2022 in central Slovenia to investigate the dominant MRSA genotypes causing bloodstream infections and to determine their antimicrobial resistance genes and virulence factors.
Materials and methods
Settings
Slovenia has an area of 20,273 km2 and a population of 2 approximately million. The Department of Bacteriology of the Institute of Microbiology and Immunology of the Faculty of Medicine, University of Ljubljana (IMI) serves three hospitals and covers a central region of about 700 000 inhabitants.
Collection of bacterial strains
All non-duplicated MRSA strains isolated from blood cultures of individual patients between January 2019 and December 2022 at IMI, were included in our retrospective study. IMI serves three hospitals and processes approximately 38 000 blood cultures per year, using the BD Bactec FX400 (BD, Sparks, USA) according to the manufacturer’s instructions. Blood cultures were incubated for 5 days, and all positive bottles were processed according to the laboratory’s standard operating procedures, which include Gram staining, subculture, identification, and antimicrobial susceptibility testing.
Identification of bacteria and phenotypic antimicrobial susceptibility testing
S. aureus isolates were identified by MALDI-TOF mass spectrometry (Bruker Daltonic GmBH, Bremen, Germany), and MRSA isolates were detected by antibiotic susceptibility testing using the agar disk diffusion method according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [18]. The antibiotics tested were penicillin, cefoxitin, gentamicin, erythromycin, clindamycin, tetracycline, ciprofloxacin, trimethoprim-sulfamethoxazole, rifampin, linezolid, and mupirocin (BD, Sparks, USA). The minimal inhibitory concentration (MIC) of vancomycin and teicoplanin was determined using the E-test (bioMerieux, Marcy-l’Etoile, France).
Patient data
For all patients with MRSA-positive blood cultures, data on gender, age, and date of sample collection were extracted from the MBL laboratory information system (SRC Infonet, Kranj).
Whole genome sequencing
Whole genome sequencing was performed on 82 isolates. DNA was extracted using the DNeasy blood and tissue kit (Qiagen), and genomic libraries were prepared with Nextera XT Library Preparation Kit (Illumina). Isolates were sequenced, targeting a minimum coverage of 150x with the NextSeq 550 System (Illumina) using 2 × 149 bp paired-end reads chemistry. Fastp v0.23.2 [19] was used to trim the raw reads of adapter sequences and low quality reads. Quality of both raw and trimmed reads was assessed with FastQC v0.11.9 [20]. Assembly of trimmed reads into contigs was done with SPAdes v3.15.3 [21] using the default kmer values and “--careful” parameters. Quast v5.2.0 [22] was used for quality assessment of the assemblies.
MLST, cgMLST, spa and SCCmec typing
Sequence types of isolates were determined in silico using the S. aureus typing database on PubMLST [23]. SpaTyper v0.3.3 [24] was used for typing of isolates, using the repeat sequences and repeat orders available on the spa typing website (https://spa.ridom.de/index.shtml) [25]. Staphylococcal chromosomal cassette typing was performed using SCCmecFinder v1.2 [26] with default parameters. Phylogenetic analysis of isolates was performed with core genome MLST, using the typing scheme for S. aureus available in Ridom SeqSphere+ [27]. A neighbor joining tree (NJT) was constructed based on the cgMLST allele differences, ignoring pairwise missing values and using only samples with more than 95% of the 1861 available alleles. The iTOL online phylogenetic tree display tool was used to visualize and annotate the tree [28].
Virulence factors and antimicrobial resistance genes
Antimicrobial resistance genes present in isolates were identified with ResFinder 4.1, using default parameters [29]. Virulence factors were identified with ABricate v1.0.1 [30] with default settings, using the virulence factor database (VFDB) [31], as well as the AlereMicroarray_Virulence tool available in Ridom SeqSphere + software [32].
Data availability
The generated raw reads of all isolates were submitted to the European nucleotide archive under the study accession number PRJEB66124.
Results
Between 2019 and 2022, a total of 1023 nonrepeating S. aureus isolates were identified in patients with bloodstream infections. Eighty-two (8.0%) were characterized as MRSA and included in our study.
Molecular epidemiology
All identified MRSA carried the mecA gene; none of the isolates carried the mecC gene. The MRSA blood culture isolates belonged to six different clonal complexes (CC) and to eight different sequence types (ST). The most prevalent CC was CC5 (64.7%, n = 53), followed by CC97 (18.3%, n = 15), CC22 (9.8%, n = 8), CC1 (2.4%, n = 2), CC45 (2.4%, n = 2) and CC398 (2.4%, n = 2). The most prevalent ST was ST225 (45.1%, n = 37), followed by ST97 (18.3%, n = 15), ST2883 (15.9%, n = 13), ST22 (9.8%, n = 8), ST5 (3.7%, n = 3), ST1 (2.4%, n = 2), ST45 (2.4%, n = 2) and ST 398 (2.4%, n = 2). Genetic diversity of MRSA isolates was determined using spa typing. A total of 16 spa types were observed, and the most predominant type was t003 (31.7%, n = 26). Two isolates were non-typable by spa typing. In SCCmec typing, three types (II, IV and V) were determined among the 82 MRSA isolates. The most frequently confirmed type was SCCmec II (63.4%, n = 52), followed by SCCmec IV (34.1%, n = 28) and SCCmec V (2.5%, n = 2). With agr typing, three types (I, II and III) were determined. 32.9% (n = 27) MRSA carried agr I, 64.6% (n = 53) carried agr II in and 2.5% (n = 2) agr III.
Phenotypic and genotypic antimicrobial resistance
All 82 MRSA isolates carried the mecA gene and were phenotypically resistant to cefoxitin. Antimicrobial susceptibility results showed that all isolates were fully susceptible to several antibiotics tested: linezolid, rifampicin, vancomycin, teicoplanin and trimethoprim-sulfamethoxazole.
91.5% of isolates (n = 75) carried the blaZ gene encoding a penicillinase, while 8.5% (n = 7) of MRSA isolates, belonging to ST225 (t003, n = 4), ST2883 (t4336, n = 2) and ST398 (t011, n = 1) did not have the blaZ gene and were phenotypically resistant to penicillin.
Among macrolides and lincosamides, 74.4% (n = 61) of MRSA isolates showed resistance to erythromycin and clindamycin. Of these, 82% (n = 50) had the constitutive erythromycin-clindamycin resistance phenotype, while 18% (n = 11) had the inducible erythromycin-clindamycin resistance phenotype. Fifty-nine (72%) isolates were resistant to clindamycin, ciprofloxacin, and erythromycin, most of which belonged to ST225 (n = 36) and ST2883 (n = 13).
9.1% (n = 7) of MRSA isolates belonging to ST1 (t127, n = 2), ST45 (t6890, n = 2), ST225 (t003, n = 1) and ST398 (t011, n = 2) were phenotypically resistant to tetracycline. One isolate that belonged to ST225 (t003) lacked antibiotic resistance genes tetK and tetM and did not correlate with the phenotypic results.
For the aminoglycosides, only 1.3% (n = 1; t359, ST97) of isolates showed phenotypic resistance to gentamicin, while the resistant gene aadD (n = 2; t002, ST5), ant(6)-Ia (n = 1; t127, ST1), ant(9)-Ia (n = 52; t002 (n = 2), ST5; t003 (n = 26), t014 (n = 4), t045 (n = 5), t1227 (n = 1), NT (n = 1), ST225; t4336 (n = 12), t19843 (n = 1), ST2883 and aph (3´)-III (n = 1, t127, ST1) were determined. Agreement between phenotypic and genotypic susceptibility testing is shown in Table 1.
Virulence genes
Virulence genes were diverse and correlated with typing characteristics (Fig. 1). Genes encoding aureolysins (aur), alpha (hla), beta (hlb), gamma-hemolysin A, B, C (hlgABC) and leukocidins (lukD, lukE), were detected at frequencies of 100%, 100%, 100%, 100%, and 85.4% of all MRSA isolates, respectively. Staphylococcus complement inhibitor (scn) and staphylokinases (sak) were detected in 96.3% of all MRSA isolates, whereas no virulence genes were confirmed in three MRSA isolates belonging to ST97 (t359, n = 2) and ST398 (t011, n = 1). 95.1% MRSA isolates contained the cap5 gene, while the cap8 gene carried 4 isolates that belonged to ST1 (n = 2, spa type t127) and ST45 (n = 2, spa type t6890). The intracellular adhesion gene icaA was present in all isolates.
None of the isolates carried Panton-Valentine leukocidin (lukSF-PV), exfoliative toxins (eta, etb, etd) or toxic shock toxin (tst). Many of the isolates carried an enterotoxin or enterotoxin-like genes, including sec (n = 2), sed (n = 48), seg (n = 63), seh (n = 2), sei (n = 63), sej (n = 48), sel (n = 2), sem (n = 63), sen (n = 63), seo (n = 63), sep (n = 46), ser (n = 48), and seu (n = 62). The majority of isolates (79.3%, n = 65) carried ≥ 1 enterotoxin gene, while 17 isolates had no enterotoxin genes. These isolates belonged to ST97, spa type t359 (n = 13), t2770 (n = 1), NT (n = 1) and ST398, spa type t011 (n = 2).
Discussion
In our study, we phenotypically and genotypically analysed 82 MRSA strains isolated from patients with bloodstream infections in a central region of Slovenia in 2019–2022. This represents approximately one third of Slovenian MRSA isolates submitted to EARS-Net.
In our study, three SCCmec types (II, IV, V), eight ST types (ST1, ST5, ST22, ST45, ST97, ST225, ST398, ST2883), six CC complexes (CC1, CC5, CC22, CC45, CC97, CC398) and 16 spa types (t002, t003, t010, t011, t014, t022, t045, t127, t359, t790, t1227, t2770, t4336, t6890, t11581, t19843) were identified.
According to SKUOPZ data, MRSA isolates from clinical samples in Slovenia in 2017 were resistant to erythromycin, clindamycin, and ciprofloxacin in 75.2%, 74.3%, and 73%, respectively [10]. In our study we show that the majority of MRSA strains (n = 59) were also resistant to multiple antimicrobials (erythromycin, clindamycin, and ciprofloxacin) and belonged to ST225 (n = 36), ST2883 (n = 13), ST22 (n = 7), ST5 (n = 2) and ST97 (n = 1). MRSA strains (n = 20) that were susceptible to erythromycin, clindamycin and ciprofloxacin belonged to ST1 (n = 1), ST5 (n = 1), ST22 (n = 1), ST45 (n = 2), ST97 (n = 13), and ST398 (n = 2). The concordance of phenotype and genotype with the identified resistance gene was 97.8%. Discordant results for penicillin resistance could be due to incorrect reading of the edge of the inhibition zone as fuzzy or sharp according to EUCAST standards [18]. Discrepancies between phenotype and genotype resistance could either be due to the isolates having a nonexpressed resistance gene, plasmid loss, the isolates lacking the resistance gene or conferring other resistance mechanisms. All isolates of our most prevalent ST225-II and ST2883-II carried resistance genes against erythromycin and exhibited phenotypic resistance. Macrolide, ciprofloxacin, and mupirocin resistance phenotypes were described as biomarkers of epidemiological success in a recent study by Baede et al., whereas gentamicin resistance was associated with sporadicity [33]. These observations are consistent with our results.
CC5 (n = 53, 64.7%) was the predominant clone identified among MRSA strains and is the most prevalent clone causing HA-MRSA infections in the Western hemisphere [34]. In our study, CC5 included ST5, ST225 and ST2883.
ST225-II, also known as the Rhine Hesse MRSA clone [35] was identified in 37 isolates (45.1%). The most common spa type in our study that belonged to ST225-II, t003 (n = 26), was also one of the most prevalent spa types causing invasive MRSA infections in Europe [7]. According to the study by Kotnik-Kervokijan et al. [13], these spa types t003 have been found in Slovenia previously, so ST225-II clone is probably one of the endemic clones circulating in our country. In our study, all ST225-II strains carried inducible resistance by the erythromycin ermA gene and the aminoglycoside resistance gene ant(9)-Ia, as well as the enterotoxin genes seg, sei, sem, sen and seo.
ST2883-II was identified in 13 isolates (15.9%), twelve of which belonged to spa type t4336, and one to t19843. A novel ST2883 was identified among suspected CA-MRSA in Slovenia in a previous study in 2010 [12]. To our knowledge, no further data are available. All ST2883-II MRSA isolates were resistant to penicillin, erythromycin, clindamycin, and ciprofloxacin, carried lukED genes, enterotoxin genes (seg, sei, sem, sen, seo, seu), and haemolysin genes (alpha, beta, gamma). Alleles of sequence types ST225 and ST2883 only differ in a single nucleotide in the gene yqiL. Based on pairwise allele differences, some of the isolates belonging to ST225 are more closely related to ST2883 than other isolates belonging to the same ST. The two sequence types are highly similar; of the seven genes used for typing, there is a single nucleotide difference in the yqiL gene. ST2883 clone is a novel variant of ST225 and an important clone circulating in Slovenia that causes bloodstream infections.
ST5-II was detected in two isolates (2.4%, spa type t002) and is a pandemic strain also known as the New York/Japan clone [35]. In 2011, spa type t002 was the fourth most common spa type among invasive MRSA in Europe according to Grundmann et al. [7]. Both strains harboured the macrolide resistance gene ermA, aminoglycoside resistance gene aadD and the virulence-associated genes sed, seg, sei, sem, sen, seo, ser, and seu. We also detected one (1.3%, t010) MRSA strain belonging to ST5-IV, also known as the Paediatric clone [35]. The strain was resistant to penicillin and carried the blaZ gene, the haemolysin genes (alpha, beta, gamma), the lukED gene, and- the enterotoxin genes (sed, seg, sei, sej, sem, sen, seo, ser, and seu).
The second most frequent clone was CC97, ST97-IVc, which was identified in 15 isolates (18.3%). Of these, 13 belonged to spa type t359, one to spa type t2770 and one was non-typable. This clone is usually associated with livestock and rarely with human infections [36, 37]. Spa type t359 was previously confirmed in Slovenia among suspected human CA-MRSA isolates in 2014 and 2015 [11]. ST97-IV-t359 has also been found in Bosnia and Herzegovina and in a maternity hospital in Ireland [38, 39]. Although reports of ST97-IV clone are less common in Europe, it is considered an important pathogen in Slovenia. In our study, all ST97 MRSA isolates were resistant to penicillin and cefoxitin and carried the lukED gene. The ST97 isolates were negative for exfoliative toxin genes, toxic shock syndrome toxin genes and enterotoxins genes, similarly to isolates of other major LA-MRSA clones (i.e., CC398 and CC1) [2]. Two ST97 isolates were sak and scn genes negative, indicating a possible animal origin [1, 2].
The third most frequent clone CC22, ST22-IV was identified in eight isolates (9.8%), of which three belonged to spa type t022, four to t790 and one to t11581. ST22-IV is a pandemic strain also known as EMRSA-15 [35]. According to the study by Grundmann et al. the spa type t022 was the ninth most common spa type among invasive MRSA in Europe in 2011 [7]. All isolates belonging to ST22-IV in our study were resistant to penicillin, 25% to ciprofloxacin, erythromycin and clindamycin, and harboured haemolysin (alpha, beta, gamma) and enterotoxin genes (seg, sei, sem, sen, seo, and seu).
We have also identified other clones associated with LA-MRSA: two isolates belonged to ST1-IVa (2.4%, spa type t127) and two belonged to ST398-V (2.4%, spa type t011). All strains were resistant to penicillin, tetracycline, 25% to erythromycin and clindamycin. spa type t127 carried the lukED gene and the enterotoxin gene sek. The sak and the scn genes were negative in one strain, spa type t011, ST398, indicating a possible animal origin [1, 2]. None of the ST398-V-t011 strains had genes associated with virulence (enterotoxins, Pantone-Valentine leukocidin). According to our previous study among suspected CA-MRSA [11, 12] and the Kotnik-Kervokijan study [13], the ST398 clone has spread in the community and healthcare settings throughout the country. Currently, the number of severe infections caused by ST398 remains low in a central region of Slovenia. Larsen and colleagues reported an increased number of LA-MRSA ST398 bacteremias in Denmark, where most patients lived in rural areas but had no contact with livestock. The researchers predicted that the number of severe infections and deaths will increase if LA-MRSA ST398 is allowed to spread in the general population [39].
The final CC45 clone, ST45-IV was identified in two isolates (2.4%, spa type t6890) [35]. Both strains were resistant to penicillin and tetracycline, and they carried haemolysin (alpha, beta, gamma) and enterotoxin genes (sec, e.g., sei, sel, sem, sen, seo, and seu).
Surprisingly, we did not find any mecC positive clone, otherwise found in previous studies, in which mecC positive strains were detected both in asymptomatic carriers and also in clinical samples (sputum, blood culture) [11, 12, 14].
The virulence factors of S. aureus play an important role during pathogenesis [1,2,3]. Overall, in our study, all MRSA isolates harboured exotoxin genes (hla, hlb and hlg), adhesin (icaA) and enzymes (sspA, sspB) which may facilitate invasion and immune evasion. We also confirmed that over 50% of bloodstream MRSA isolates encode seg, sei, sem, sen, seo and / or seu [40]. Surprisingly, all isolates that belonged to ST97 had none of the enterotoxin genes, while two isolates that belonged to ST1 harboured only seh. According to the Bennett and Thomsen study the leucocidin toxin genes lukED are common across S. aureus lineages, with approximately 87% clinical isolates carrying lukED [41]. A similar percentage (85.4%) was found in our study, and only MRSA strains that belonged to ST22, ST45 and ST398 were lukED negative.
The main limitation of our study is the incomplete access to medical data and data on possible patient transfers between hospitals. Therefore, we could not investigate possible transmission events or investigate correlation over time. Additionally, two isolates in our study were non-typeable by spa typing. Due to limitations of short-read sequencing when it comes to repetitive regions, as well as possible low sequencing coverage in this region, spa types could not be reliably determined, because multiple spa repeats were assembled, or the 5’ or 3’ signatures were missing. Spa types were considered reliable when only one repeat sequence was recognised and the 5’ and 3’ signatures were found in the correct positions, as was the case for other isolates. For further studies, long-read sequencing could be employed to resolve these limitations. Like other epidemiologic studies, our study is also limited by the selected isolates.
Conclusions
In summary, we present diverse lineages of MRSA causing bacteraemia in a central region of Slovenia. Currently, the new genetic MRSA lineage ST2883 plays an important role in the spread in the Slovenian population. Clonal replacement is frequent, and the identification of emerging lineages should be monitored in the future to detect changes in the molecular epidemiology of MRSA and guide interventions to reduce the burden of MRSA in Slovenia.
Data availability
The generated raw reads of all isolates were submitted to the European nucleotide archive under the study accession number PRJEB66124.
References
Lee AS, de Lencastre H, Garau J, Kluytmans J, Malhotra-Kumar S, Peschel A, Harbarth S (2018) Methicillin-resistant Staphylococcus aureus. Nat Rev Dis Primer 31:18033. https://doi.org/10.1038/nrdp.2018.33
Bal AM, Coombs GW, Holden MTG, Lindsay JA, Nimmo GR, Tattevin P, Skov RL (2016) Genomic insights into the emergence and spread of international clones of healthcare-, community- and livestock-associated meticillin-resistant Staphylococcus aureus: blurring of the traditional definitions. J Glob Antimicrob Resist 6:95–101. https://doi.org/10.1016/j.jgar.2016.04.004
Recker M, Laabei M, Toleman MS et al (2017) Clonal differences in Staphylococcus aureus bacteraemia-associated mortality. Nat Microbiol 2:1381–1388. https://doi.org/10.1038/s41564-017-0001-x
Köck R, Becker K, Cookson B et al (2010) Methicillin-resistant Staphylococcus aureus (MRSA): burden of disease and control challenges in Europe. Euro Surveill 15:19688. https://doi.org/10.2807/ese.15.41.19688-en
Tenover FC, Tickler IA, Goering RV, Kreiswirth BN, Mediavilla JR, Persing DH, MRSA Consortium (2012) Characterization of nasal and blood culture isolates of methicillin-resistant Staphylococcus aureus from patients in United States hospitals. Antimicrob Agents Chemother 56:1324–1330. https://doi.org/10.1128/AAC.05804-11
Johnson AP (2011) Methicillin-resistant Staphylococcus aureus: the European landscape. J Antimicrob Chemother 66:iv43–iv48. https://doi.org/10.1093/jac/dkr076
Grundmann H, Schouls LM, Aanensen DM et al (2014) The dynamic changes of dominant clones of Staphylococcus aureus causing bloodstream infections in the European region: results of a second structured survey. Euro Surveill 19:20987. https://doi.org/10.2807/1560-7917.es2014.19.49.20987
Gagliotti C, Högberg LD, Billström H et al (2021) Staphylococcus aureus bloodstream infections: diverging trends of meticillin-resistant and meticillin-susceptible isolates, EU/EEA, 2005 to 2018. Euro Surveill 26:2002094. https://doi.org/10.2807/1560-7917.ES.2021.26.46.2002094
European Centre for Disease Prevention and Control (2022) Antimicrobial resistance in the EU/EEA (EARS-Net) - Annual Epidemiological Report 2021. ECDC. https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2021. Accessed 21 Sep 2023
Štrumbelj I, Pirš M, Berce I et al (2018) Pregled občutljivosti bakterij za antibiotike - Slovenija 2017. [Annual report on Antimicrobial Susceptibility Testing - Slovenia 2017]. Ljubljana: Slovenian National Antimicrobial Susceptibility Testing Committee (SKUOPZ). https://www.dlib.si/details/URN:NBN:SI:DOC-JJI1P2FH 1. Accessed 21 Sep 2023
Dermota U, Košnik IG, Janežič S, Rupnik M (2020) Changing epidemiology of Presumptive Community-associated-methicillin-resistant Staphylococcus aureus in Slovenia in 2014–2015 compared to 2010. Zdr Varst 59:236–244. https://doi.org/10.2478/sjph-2020-0030
Dermota U, Mueller-Premru M, Švent-Kučina N et al (2015) Survey of community-associated-methicillin-resistant Staphylococcus aureus in Slovenia: identification of community-associated and livestock-associated clones. Int J Med Microbiol 305:505–510. https://doi.org/10.1016/j.ijmm.2015.05.002
Kevorkijan BK, Petrovič Ž, Kocuvan A, Rupnik M (2019) MRSA diversity and the emergence of LA-MRSA in a large teaching hospital in Slovenia. Acta Microbiol Immunol Hung 66:23546. https://doi.org/10.1556/030.65.2018.052
Dermota U, Zdovc I, Strumbelj I et al (2015) Detection of methicillin-resistant Staphylococcus aureus carrying the mecC gene in human samples in Slovenia. Epidemiol Infect 143:1105–1108. https://doi.org/10.1017/S0950268814001861
Svent-Kucina N, Pirs M, Kofol R, Blagus R, Smrke DM, Bilban M, Seme K (2016) Molecular characterization of Staphylococcus aureus isolates from skin and soft tissue infections samples and healthy carriers in the Central Slovenia region. APMIS 124:309–318. https://doi.org/10.1111/apm.12509
Cvitković Špik V, Grmek Košnik I, Lorenčič Robnik S et al (2009) Genetic characterization of methicillin-susceptible and methicillin-resistant Staphylococcus aureus strains isolated from bloodstream infections in Slovene hospitals using spa typing. Zdr Varst 48:78–84. https://doi.org/
Grundmann H, Aanensen DM, van den Wijngaard CC, Spratt BG, Harmsen D, Friedrich AW, European Staphylococcal Reference Laboratory Working Group (2010) Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med 7:e1000215. https://doi.org/10.1371/journal.pmed.1000215
The European Committee on Antimicrobial Susceptibility Testing (2020) Breakpoint tables for interpretation of MICs and zone diameters, version 10.0. https://www.eucast.org/clinical_breakpoints. Accessed 21 Sep 2023
Chen S, Zhou Y, Chen Y, Gu J (2018) Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–890. https://doi.org/10.1093/bioinformatics/bty560
Andrews S (2010) FastQC: A quality control tool for high throughput sequence data. Babraham Bioinformatics. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed 21 Sep 2023
Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A (2020) Using SPAdes de novo assembler. Curr Protoc Bioinf 70:e102. https://doi.org/10.1002/cpbi.102
Mikheenko A, Prjibelski A, Saveliev V, Antipov D, Gurevich A (2018) Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 34:142–150. https://doi.org/10.1093/bioinformatics/bty266
Jolley KA, Bray JE, Maiden MCJ (2018) Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. https://doi.org/10.12688/wellcomeopenres.14826.1
Sanchez-Herrero JF (2020) spaTyper: Staphylococcal protein A (spa) characterization pipeline (v0.3.1). Zenodo. https://zenodo.org/record/4063625. Accessed 21 Sep 2023
Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, Vogel U (2003) Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol 41:5442–5448. https://doi.org/10.1128/JCM.41.12.5442-5448.2003
Kaya H, Hasman H, Larsen J et al (2018) SCCmecFinder, a web-based tool for typing of staphylococcal cassette chromosome mec in Staphylococcus aureus using whole-genome sequence data. mSphere 3:e00612-17. https://doi.org/10.1128/mSphere.00612-17
Leopold SR, Goering RV, Witten A, Harmsen D, Mellmann A (2014) Bacterial whole-genome sequencing revisited: portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol 52:2365–2370. https://doi.org/10.1128/JCM.00262-14
Letunic I, Bork P (2021) Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–296. https://doi.org/10.1093/nar/gkab301
Bortolaia V, Kaas RS, Ruppe E et al (2020) ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother 75:3491–3500. https://doi.org/10.1093/jac/dkaa345
Seemann T, Abricate (2023) Sep. Github. https://github.com/tseemann/abricate Accessed 21
Chen L, Zheng D, Liu B, Yang J, Jin Q (2016) Nucleic Acids Res 44:D694–697. https://doi.org/10.1093/nar/gkv1239. VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on
Strauß L, Ruffing U, Abdulla S et al (2016) Detecting Staphylococcus aureus virulence and resistance genes: a comparison of whole-genome sequencing and DNA microarray technology. J Clin Microbiol 54:1008–1016. https://doi.org/10.1128/JCM.03022-15
Baede VO, Gupta A, Knight GM et al (2023) Markers of epidemiological success of methicillin-resistant Staphylococcus aureus isolates in European populations. Clin Microbiol Infect 29:1166–1173. https://doi.org/10.1016/j.cmi.2023.05.015
Challagundla L, Reyes J, Rafiqullah I et al (2018) Phylogenomic classification and the evolution of clonal complex 5 methicillin-resistant Staphylococcus aureus in the western hemisphere. Front Microbiol 9:1901. https://doi.org/10.3389/fmicb.2018.01901
Monecke S, Coombs G, Shore AC et al (2011) A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE 6:e17936. https://doi.org/10.1371/journal.pone.0017936
Silva V, Araújo S, Monteiro A et al (2023) Staphylococcus aureus and MRSA in livestock: antimicrobial resistance and genetic lineages. Microorganisms 11:124. https://doi.org/10.3390/microorganisms11010124
Rao S, Linke L, Magnuson R, Jauch L, Hyatt DR (2022) Antimicrobial resistance and genetic diversity of Staphylococcus aureus collected from livestock, poultry and humans. One Health Amst Neth 15:100407. https://doi.org/10.1016/j.onehlt.2022.100407
Numanović F, Dermota U, Smajlović J et al (2021) Characterization and clonal representation of MRSA strains in Tuzla Canton, Bosnia and Herzegovina, from 2009 to 2017. Med Glas (Zenica) 18:38–46. https://doi.org/10.17392/1265-21
Broderick D, Brennan GI, Drew RJ, O’Connell B (2021) Epidemiological typing of methicillin resistant Staphylococcus aureus recovered from patients attending a maternity hospital in Ireland 2014–2019. Infect Prev Pract 3:100124. https://doi.org/10.1016/j.infpip.2021.100124
Belkum A, Melles DC, Snijders SV et al (2006) Clonal distribution and differential occurence of the enterotoxin gene cluster, egc, in carriage-versus bacteremia-associated isolates of Staphylococcus aureus. J Clin Microbiol 44:1555–1557. https://doi.org/10.1128/JCM.44.4.1555-1557.2006
Bennett MR, Thomsen IP Epidemiological and clinical evidence for the role of toxin in S. Aureus human disease. Toxins 12:408. https://doi.org/10.3390/toxins12060408
Funding
This work was supported by the Slovenian Research Agency (grant numbers P3-0083).
Author information
Authors and Affiliations
Contributions
UD, TT and IV conceived the study and provided the required resources. KSS and ACS performed the practical work and the bioinformatics analysis, UD performed the remaining data analysis. UD, ACS and IV wrote the manuscript. KSS and TT critically reviewed and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval
This study was performed in accordance with the Declaration of Helsinki. The samples used in this study were collected during routine patient management without the need for additional sampling. As such, this study did not require ethics committee approval or informed consent from the patients.
Consent to participate
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dermota, U., Šturm, A.C., Triglav, T. et al. Whole genome sequencing and molecular epidemiology of methicillin-resistant Staphylococcus aureus isolated from patients with bacteraemia in Slovenia. Eur J Clin Microbiol Infect Dis 43, 969–977 (2024). https://doi.org/10.1007/s10096-024-04802-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-024-04802-1