Abstract
Rapid detection of pathogens causing bloodstream infections (BSI) directly from positive blood cultures is of highest importance in order to enable an adequate and timely antimicrobial therapy. In this study, the utility and performance of a recently launched next-generation fully automated test system, the Biofire FilmArray® Blood Culture Identification 2 (BCID2) panel, was evaluated using a set of 103 well-characterized microbial isolates including 29 antimicrobial resistance genes and 80 signal-positive and 23 signal-negative clinical blood culture samples. The results were compared to culture-based reference methods, MALDI-TOF, and/or 16S rDNA sequencing. Of the clinical blood culture samples, 68 were monomicrobial (85.0%) and 12 polymicrobial (15.0%). Six samples contained ESBL (blaCTX-M), two MRSA (mecA), and three MRSE (mecA) isolates. In overall, the FilmArray BCID2 panel detected well on-panel targets and resistance markers from mono- and polymicrobial samples. However, one Klebsiella aerogenes and one Bacteroides ovatus were undetected, and the assay falsely reported one Shigella flexneri as Escherichia coli. Hence, the sensitivity and specificity for detecting microbial species were 98.8% (95%CI, 95.8–99.9%) and 99.9% (95%CI, 99.8–99.9%), respectively. The sensitivity and specificity for detecting of resistance gene markers were 100%. The results were available within 70 min from signal-positive blood cultures with minimal hands-on time. In conclusion, the BCID2 test allows reliable and simplified detection of a vast variety of clinically relevant microbes causing BSI and the most common antimicrobial resistance markers present among these isolates.
Similar content being viewed by others
Introduction
High mortality and morbidity rates are associated with bloodstream infections (BSIs) [1, 2]. In polymicrobial BSIs, which are not uncommon (ranging from 10 to 20% of all diagnosed BSIs), the mortality rates are even higher than with monomicrobial BSIs [3,4,5,6]. Therefore, rapid administration of effective antimicrobial treatment is crucial for patient survival [7, 8]. To support this, timely and accurate identification of microorganisms are central to the optimal management of BSIs.
The “gold standard” for screening of BSIs is still blood culture followed by Gram-staining and conventional culture-based techniques. The conventional techniques have the advantage of providing accurate species identification and antimicrobial susceptibility profile but the disadvantage of requiring 1–2 additional days after signal-positive blood cultures. During the last few years, various methods have been developed to optimize the detection of the etiological agent of BSI such as fluorescence hybridization probes [9,10,11], pathogenic-specific PCR [12, 13], and MALDI-TOF MS [14,15,16]. Despite the advantages of these approaches, many of them, however, comprise multiple assay steps for the purification and extraction of the microbes and their target protein or DNA. Moreover, most of them provide limited information from polymicrobial blood culture samples. Modern automatization and advances in molecular technology, however, have recently led to the development of molecular-based high multiplex platforms, such as the FilmArray (bioMérieux, France), the ePlex (GenMark Diagnostics, USA), and the Verigene (Luminex, USA), for direct and rapid multimicrobial detection from signal-positive blood culture bottles [17,18,19]. These assays allow significantly shorter time to detection than conventional methods and a high performance even with polymicrobial samples, although some limitations in the first-generation assay panel content were evident (e.g., absence of the most common resistance genes from the panel) [17,18,19].
Here, the performance and utility of the recently launched updated version of the FilmArray Blood Culture Identification 2 panel, the BCID2 assay (bioMérieux, France), were evaluated. According to the manufacturer, the BCID2 panel identifies simultaneously 33 species/genus targets and 10 antimicrobial resistance genes. This is nine microbial targets and seven genes coding for antimicrobial resistance more than with the first-generation BCID [20] assay.
Materials and methods
Simulated blood culture samples
The ability of the FilmArray BCID2 panel to detect microbes from mono- and polymicrobial samples was investigated using a set of bacterial (n = 83) and fungal (n = 20) isolates originating from either ATCC or clinical blood culture specimens (Table 1). Among the bacterial isolates, 29 resistance marker genes, relevant for this study, were also included. All culture collection isolates from clinical origin had been well-identified by MALDI-TOF (Vitek MS, bioMérieux, France) and/or by 16S rDNA sequencing [21]. Samples of simulated signal-positive blood cultures were prepared by mixing 3 mL of whole blood and 8 mL of blood culture bottle medium (BacT/Alert, bioMérieux) and spiked with culture collection isolate(s) in a concentration of approximately 3 × 106 CFU/ml. In total of 21 samples were prepared, of which 17 were pooled (polymicrobial), containing up to eight microbes per sample, and four contained one microbe only (Table 2).
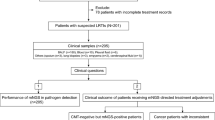
Clinical samples
The clinical blood culture samples were collected at the region of HUS hospital district and analyzed at HUS Diagnostic Center, Department of Clinical Microbiology (HUSLAB, Finland). In HUSLAB, approximately 140 000 blood culture specimens are screened each year, of which approximately 7% are growth-positive. E. coli and S. aureus among few other common bacteria causing BSI form vast majority of strains isolated from positive blood cultures samples. Testing consecutive samples in this setting would yield, with high probability, a one-sided view of the test in hand given the limited amount of test kits assigned for the study. To overcome these limitations and to simulate situations where the results would yield maximal clinical impact following criteria for the samples to be tested were made. A single positive blood culture bottle per patient would be tested if a) small gram-positive rod, gram-negative cocci, yeast, or two or more findings would be identified in gram stain; and/or b) the patient was known to be a carrier of ESBL producing Enterobacteriaceae and/or MRSA. In addition, to provide a larger spectrum of causative agents to be tested and to ensure that all of the test kits assigned for the study would be used in limited time some of the samples were selected by authors guided by the principle to provide a diverse sample material for the study. A total of 103 blood culture bottles (BacT/Alert, bioMérieux, France), one bottle per patient, confirmed to contain either Gram-negative bacteria, Gram-positive rods, Gram-positive diplococci, cocci in chains, yeast, or polymicrobial growth by Gram-staining and evaluation by a clinical microbiologist, were analyzed between November 2020 and January 2021 with the FilmArray BCID2 assay and routine reference methods. Routine methods included subculture from the signal-positive bottle on rich universal agars, such as blood, chocolate, and fastidious anaerobe agar plates with incubation at 35 °C in normal atmosphere as well as in elevated CO2 and in anaerobic conditions for 24 to 48 h. The short-incubation MALDI-TOF (si-MALDI-TOF) technique [21] was used for the preliminary identification of microbial growth from the secondary cultures after 6 hours of incubation. The identifications were confirmed by repeating the MALDI-TOF (Vitek MS, bioMérieux, France) analysis after 24 or 48 hours of incubation from distinct colonies. In case of unclear results, the confirmation was done by 16S rDNA sequencing [21]. Antimicrobial susceptibilities were determined using disc (Oxoid, UK) and/or MIC-gradient (M.I.C.E, Oxoid, Basingstoke, UK and ETest, bioMérieux, France) diffusion tests on Mueller-Hinton agar (BD, USA) at 35 °C for 24 to 48 hours. Phenotypic susceptibility profile for each isolate was interpreted according to the EUCAST standard [22]. Genotypic confirmation was done in national reference laboratory (Finnish Institute for Health and Welfare).
Sample runs with the FilmArray BCID2 assay were performed according to the manufacturer’s instructions. Performance of the BCID2 panel was calculated using MedCalc statistical analysis software (MedCalc Software Ltd, Belgium). The 95% confidence intervals (95% CI) were approximated using the Clopper-Pearson method, i.e., binomial exact confidence interval method.
The Fisher’s exact test was used to determine statistical significance of the differences among the BCID2 panel and routinely used reference methods in species and resistance marker identification within specified time period (within 8 hours from signal of positivity).
Results
From the simulated sample mixes, the BCID2 panel provided a correct identification in 99.9% (95% CI, 99.7–100%) of all target microbes and in 100% (95% CI, 98.7–100%) of all resistance marker genes (Table 1). The false detections consisted of one Klebsiella aerogenes isolate, which was repeatedly undetected from different sample mixes and Shigella flexneri, which was misidentified as Escherichia coli. In addition, Bacteroides ovatus, a member of B. fragilis group, was not detected as B. fragilis, whereas B. thetaiotaomicron, also a member of B. fragilis group, was detected as B. fragilis. This was considered as a false-negative detection for the B. ovatus isolate. In general, the BCID2 panel showed excellent performance from the multipositive samples providing correct identification even from samples containing 11 different targets.
Of the 103 clinical blood culture samples, 80 were signal-positive containing either Gram-negative bacteria, Gram-positive rods, diplococci, cocci in chains, yeasts, or polymicrobial growth by Gram-staining. The proportion of polymicrobial samples was 15.0% (n = 12). The remaining 23 samples, included in the study, were signal- and growth-negative. In total, 36 Gram-positive bacteria, consisting of 12 different species, 36 Gram-negative bacteria, consisting of nine different species, and five yeasts, consisting of two different species, were correctly identified by the BCID2 panel (Table 3). The most common microbes recovered were E. coli (n = 15), Streptococcus spp. (n = 11), Klebsiella spp. (n = 10), and L. monocytogenes (n = 6). The BCID2 panel covered 81.9% of the microbes found from the blood culture samples. The missing (off-panel) species were mainly anaerobic Gram-negative rods and aerobic Gram-positive rods other than Listeria spp. One unresolved discordant result was observed from a sample containing Enterococcus avium, Pseudomonas aeruginosa, and E. coli from where the BCID2 panel yielded Streptococcus spp., P. aeruginosa, and E. coli positive result. Growth appropriate for streptococci was not observed on any agar plates nor was the isolate confirmed by any other method. Along with the Gram-negative and Gram-positive bacteria, 10 resistance genes (six blaCTX-M and four mecA/C genes) were detected and correctly identified by the BCID2 panel (Table 3). No additional resistance markers were found by the routine reference methods. Thus, the overall sensitivity and specificity of the BCID2 panel from the clinical sample set were 100% (95% CI, 95.9–100%) and 99.9% (95% CI, 99.8–100%), respectively.
Species identification with the routine culture-based methods was available for 68.0% of the samples within 24–48 h, when short-incubation (si-)MALDI-TOF could not be utilized (e.g., with polymicrobial growth and slowly growing organism), and for 32.0% of the samples within 6–8 h, when si-MALDI-TOF could be utilized. For antimicrobial susceptibilities, the total turnaround time was 24–48 h with the routine methods. The direct analysis of the clinical blood culture samples with the automated BCID2 panel enabled species identification and detection of the most common antimicrobial resistance genes within 70 min. This provided significant (P value < 0.0001) time reduction as compared with the routine methods.
The BCID2 panel invalidity rate from the clinical blood culture bottle samples was 1.0% (1/103).
Discussion
In the current study, the performance and usability of the FilmArray BCID2 panel for rapid detection of organism from blood culture samples were evaluated. The new BCID2 panel has been improved greatly as it detects nine microbial targets and seven resistance genes more than the first-generation BCID panel [20, 23]. In addition, the assay run time has been reduced from 90 to 70 min. Hence, the panel provides a detection of 33 most common microbial species/genus causing BSI and 10 antimicrobial resistance marker genes reducing the time to species identification and detection of major resistance genes by at least a full day as compared with conventional culture-based methods.
The performance of the BCID2 panel was excellent providing overall 100% sensitivity and 100% specificity for on-panel antimicrobial resistance markers and 98.8% sensitivity and 99.9% specificity for on-panel microbial targets. Of the four cases from which the BCID2 panel failed to correctly identify the isolates, two contained off-panel organisms (E. avium and S. flexneri) according to reference methods. Of these, S. flexneri in monomicrobial growth was falsely identified as E. coli which is a severe misidentification, due to the low incidence but potentially life-threatening outcomes of S. flexneri bacteremia. The misidentification may occur if, e.g., β-glucuronidase gene (uidA), which is present in both species, is used for E. coli detection [24, 25]. Although these two organisms might be difficult to distinguish, there are genes that could be used for accurate differentiation and species detection [24]. Here, the S. flexneri isolate was used in simulated sample and from pure culture to ensure the result reliability.
The sample containing E. avium, however, remained unresolved as it contained polymicrobial growth of E. avium, P. aeruginosa, and E. coli of which P. aeruginosa and E. coli were correctly identified. With the reference methods, the reported Streptococcus spp. result could not be confirmed and was thus considered as a false detection from the E. avium isolate. However, since the sample contained polymicrobial growth, it is possible that streptococci, which could have been overgrown by the other species, was present in the sample but not detected by the culture-based reference methods. Sadly, the Enterococcus genus level detection, which was included in the first-generation BCID panel, has been removed from the BCID2 panel. It could have been used for the differentiation of Enterococcus spp. and Streptococcus spp. and used here to conclude whether or not the sample contained all four species (P. aeruginosa, E. coli, Enterococcus spp., and Streptococcus spp.). The differentiation of enterococci and streptococci genus from BSI samples is important, in general, due to their different natural resistance traits and thus different treatment options.
Of the other two samples yielding false-negative results with the BCID2 panel, one contained K. aerogenes (on-panel target) isolate which was confirmed as K. aerogenes by MALDI-TOF and 16S rDNA sequencing and the other one B. ovatus which is a member of the B. fragilis group. The BCID2 panel yielded no result from the B. ovatus while providing B. fragilis positive results from a sample containing B. thetaiotaomicron, another member of the B. fragilis group. All of these species are abundant in colon and important pathogens in polymicrobial infections [26]. Moreover, according to Brook, B. thetaiotaomicron and B. ovatus covers approximately 21% of all infections caused by the B. fragilis group and are associated with high mortality rate (20–30%) when inducing bacteremia [26].
Regarding the above-mentioned discrepancies it is interesting to note that three of the four misidentifications observed in this study are related to targets introduced in the panel of the new version of the kit. Consequently, further evaluation with more isolates within this group of microbes would be highly beneficial.
In addition to the FilmArray BCID2 panel, there are only few other fully automated assays that provides a broad spectrum of species identification and detection of antimicrobial resistance genes directly from signal-positive blood culture bottles. Of these, the ePlex® Blood Culture Identification (BCID) panel (Genmark Diagnostics, USA) can detect multiple microbes and resistance genes (i.e., blaKPC, blaNDM, blaVIM, blaIMP, and blaOXA-48-like genes in BCID-GN panel) within 90 min but is limited to detect only one type of organisms with a one type of test panel (e.g., BCID-GN for Gram-negative bacteria and BCID-GP for Gram-positive bacteria), and none of the panels are yet CE-IVD marked [27]. The Verigene blood culture assay (Luminex, USA) has the same limitation with test panels than the ePlex and requires approximately 2.5 h of run time per sample [28].
The limitation of this study was the low number of positive genes coding for carbapenem and vancomycin resistance in clinical samples, due to the low prevalence of carbapenem-producing organisms and vancomycin-resistant enterococci in BSI in Finland [29,30,31].
In conclusion, our results show that the FilmArray BCID2 panel proved to be a well-performing and invaluable tool for the early detection of common clinical isolates causing BSI and their antibiotic resistance genes while providing rapid aid for clinicians and helping them to administer effective antimicrobial therapy more hastily and therefore reducing patient mortality [32]. Whether to use FilmArray BCID2 panel to test all positive blood culture bottles regardless of the gram stain result or not depends on multiple factors. In an ideal situation where the most accurate and timely microbiological diagnosis is paramount FilmArray BCID2 panel could be used to test all positive blood culture bottles. However, this kind of approach can easily lead to excessive overall costs and increase the laboratory workload unnecessarily. A more frugal manner of testing only one blood culture bottle per patient might be a recommendable approach for routine diagnostic practices in most laboratories. In surroundings where resources are scarce, or the capacity of the test system is limited a more controlled approach for FilmArray BCID2 panel use could be implemented focusing on the cases for which the test provides maximal clinical impact. Before implementing this kind of diagnostic test to the routine practices, we encourage to assess the need for the test and the proposed use for the test locally with infectious disease specialists to integrate the new diagnostic means to local antimicrobial stewardship guides and to maximize the benefits of the test without increasing the overall costs of diagnostics unnecessarily. However, the overall costs of diagnostics would increase with the FilmArray BCID2 panel as compared with the conventional methods.
Data availability
All relevant data are made available in the manuscript.
Code availability
Not applicable.
Change history
08 February 2022
A Correction to this paper has been published: https://doi.org/10.1007/s10096-022-04414-7
References
Goto M, Al-Hasan MN (2013) Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin Microbiol Infect 19(6):501–509. https://doi.org/10.1111/1469-0691.12195
Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR (2001) Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med 29(7):1303–1310
Buetti N, Marschall J, Atkinson A, Kronenberg A, Swiss Centre for Antibiotic Resistance (ANRESIS) (2016) National bloodstream infection surveillance in Switzerland 2008–2014: different patterns and trends for university and community hospitals. Infect Control Hosp Epidemiol. 37(9):1060–7. https://doi.org/10.1017/ice.2016.137
Pammi M, Zhong D, Johnson Y, Revell P, Versalovic J (2014) Polymicrobial bloodstream infections in the neonatal intensive care unit are associated with increased mortality: a case-control study. BMC Infect Dis 14(14):390. https://doi.org/10.1186/1471-2334-14-390
Pavlaki M, Poulakou G, Drimousis P, Adamis G, Apostolidou E, Gatselis NK, Kritselis I, Mega A, Mylona V, Papatsoris A, Pappas A, Prekates A, Raftogiannis M, Rigaki K, Sereti K, Sinapidis D, Tsangaris I, Tzanetakou V, Veldekis D, Mandragos K, Giamarellou H, Dimopoulos G (2013) Polymicrobial bloodstream infections: epidemiology and impact on mortality. J Glob Antimicrob Resist 1(4):207–212. https://doi.org/10.1016/j.jgar.2013.06.005
Sancho S, Artero A, Zaragoza R, Camarena JJ, González R, Nogueira JM (2012) Impact of nosocomial polymicrobial bloodstream infections on the outcome in critically ill patients. Eur J Clin Microbiol Infect Dis 31(8):1791–1796. https://doi.org/10.1007/s10096-011-1503-8
Anderson DJ, Moehring RW, Sloane R, Schmader KE, Weber DJ, Fowler VG Jr, Smathers E, Sexton DJ (2014) Bloodstream infections in community hospitals in the 21st century: a multicenter cohort study. PLoS One 9(3):e91713. https://doi.org/10.1371/journal.pone.0091713
Koupetori M, Retsas T, Antonakos N, Vlachogiannis G, Perdios I, Nathanail C, Makaritsis K, Papadopoulos A, Sinapidis D, Giamarellos-Bourboulis EJ, Pneumatikos I, Gogos C, Armaganidis A, Paramythiotou E, Hellenic Sepsis Study Group (2014) Bloodstream infections and sepsis in Greece: over-time change of epidemiology and impact of de-escalation on final outcome. BMC Infect Dis. 14:272. https://doi.org/10.1186/1471-2334-14-272
Deck MK, Anderson ES, Buckner RJ, Colasante G, Davis TE, Coull JM, Crystal B, Latta PD, Fuchs M, Fuller D, Harris W, Hazen K, Klimas LL, Lindao D, Meltzer MC, Morgan M, Shepard J, Stevens S, Wu F, Fiandaca MJ (2014) Rapid detection of Enterococcus spp. direct from blood culture bottles using Enterococcus QuickFISH method: a multicenter investigation. Diagn Microbiol Infect Dis 78(4):338–42. https://doi.org/10.1016/j.diagmicrobio.2013.12.004
Stone NR, Gorton RL, Barker K, Ramnarain P, Kibbler CC (2013) Evaluation of PNA-FISH yeast traffic light for rapid identification of yeast directly from positive blood cultures and assessment of clinical impact. J Clin Microbiol 51(4):1301–1302. https://doi.org/10.1128/JCM.00028-13
Ullberg M, Özenci V (2020) Identification and antimicrobial susceptibility testing of Gram-positive and Gram-negative bacteria from positive blood cultures using the Accelerate Pheno system. Eur J Clin Microbiol Infect Dis 39(1):139–149. https://doi.org/10.1007/s10096-019-03703-y
Hirvonen JJ, von Lode P, Nevalainen M, Rantakokko-Jalava K, Kaukoranta SS (2012) One-step sample preparation of positive blood cultures for the direct detection of methicillin-sensitive and -resistant Staphylococcus aureus and methicillin-resistant coagulase-negative staphylococci within one hour using the automated GenomEra CDXTM PCR system. Eur J Clin Microbiol Infect Dis 31(10):2835–2842. https://doi.org/10.1007/s10096-012-1637-3
Tian Y, Wang L, Zhang J, Han Q, Xia XS, Song Y, Yang G (2019) CelB is a suitable marker for rapid and specific identification of Klebsiella pneumoniae by the loop-mediated isothermal amplification (LAMP) assay. Braz J Microbiol 50(4):961–967. https://doi.org/10.1007/s42770-019-00144-9
Fothergill A, Kasinathan V, Hyman J, Walsh J, Drake T, Wang YF (2013) Rapid identification of bacteria and yeasts from positive-blood-culture bottles by using a lysis-filtration method and matrix-assisted laser desorption ionization-time of flight mass spectrum analysis with the SARAMIS database. J Clin Microbiol 51(3):805–809. https://doi.org/10.1128/JCM.02326-12
Kohlmann R, Hoffmann A, Geis G, Gatermann S (2015) MALDI-TOF mass spectrometry following short incubation on a solid medium is a valuable tool for rapid pathogen identification from positive blood cultures. Int J Med Microbiol 305(4–5):469–479. https://doi.org/10.1016/j.ijmm.2015.04.004
Altun O, Botero-Kleiven S, Carlsson S, Ullberg M, Özenci V (2015) Rapid identification of bacteria from positive blood culture bottles by MALDI-TOF MS following short-term incubation on solid media. J Med Microbiol 64(11):1346–1352. https://doi.org/10.1099/jmm.0.000168
Bhatti MM, Boonlayangoor S, Beavis KG, Tesic V (2014) Evaluation of FilmArray and Verigene systems for rapid identification of positive blood cultures. J Clin Microbiol 52(9):3433–3436. https://doi.org/10.1128/JCM.01417-14
Huang TD, Melnik E, Bogaerts P, Evrard S, Glupczynski Y (2019) Evaluation of the ePlex blood culture identification panels for detection of pathogens in bloodstream infections. J Clin Microbiol 57(2):e01597-e1618. https://doi.org/10.1128/JCM.01597-18
Altun O, Almuhayawi M, Ullberg M, Ozenci V (2013) Clinical evaluation of the FilmArray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J Clin Microbiol 51(12):4130–4136. https://doi.org/10.1128/JCM.01835-13
Cortazzo V, D'Inzeo T, Giordano L, Menchinelli G, Liotti FM, Fiori B, De Maio F, Luzzaro F, Sanguinetti M, Posteraro B, Spanu T (2021) Comparing BioFire FilmArray BCID2 and BCID panels for direct detection of bacterial pathogens and antimicrobial resistance genes from positive blood cultures. J Clin Microbiol JCM.03163–20. https://doi.org/10.1128/JCM.03163-20
Halavaara M, Martelius T, Järvinen A, Antikainen J, Kuusela P, Salminen US, Anttila VJ (2019) Impact of pre-operative antimicrobial treatment on microbiological findings from endocardial specimens in infective endocarditis. Eur J Clin Microbiol Infect Dis 38(3):497–503. https://doi.org/10.1007/s10096-018-03451-5
The European Committee on Antimicrobial Susceptibility Testing (2020) Breakpoint tables for interpretation of MICs and zone diameters. Version 10.0. http://www.eucast.org. Accessed 1 Jan 2021
Kang CM, Chen XJ, Chih CC, Hsu CC, Chen PH, Lee TF, Teng LJ, Hsueh PR (2020) Rapid identification of bloodstream bacterial and fungal pathogens and their antibiotic resistance determinants from positively flagged blood cultures using the BioFire FilmArray blood culture identification panel. J Microbiol Immunol Infect S1684–1182(20):30078–30075. https://doi.org/10.1016/j.jmii.2020.03.018
Ud-Din A, Wahid S (2015) Relationship among Shigella spp. and enteroinvasive Escherichia coli (EIEC) and their differentiation. Braz J Microbiol 45(4):1131–8. https://doi.org/10.1590/s1517-83822014000400002 (eCollection 2014)
Nisa I, Qasim M, Yasin N, Ullah R, Ali A (2020) Shigella flexneri: an emerging pathogen. Folia Microbiol (Praha) 65(2):275–291. https://doi.org/10.1007/s12223-020-00773-w
Brook I (1989) Pathogenicity of the Bacteroides fragilis group. Ann Clin Lab Sci 19(5):360–376
Bryant S, Almahmoud I, Pierre I, Bardet J, Touati S, Maubon D, Cornet M, Richarme C, Maurin M, Pavese P, Caspar Y (2020) Evaluation of microbiological performance and the potential clinical impact of the ePlex ® blood culture identification panels for the rapid diagnosis of bacteremia and fungemia. Front Cell Infect Microbiol 10:594951. https://doi.org/10.3389/fcimb.2020.594951
Arroyo MA, Denys GA (2017) Parallel evaluation of the MALDI Sepsityper and Verigene BC-GN assays for rapid identification of gram-negative bacilli from positive blood cultures. J Clin Microbiol 55(9):2708–2718. https://doi.org/10.1128/JCM.00692-17
Brolund A, Lagerqvist N, Byfors S, Struelens MJ, Monnet DL, Albiger B, Kohlenberg A, European Antimicrobial Resistance Genes Surveillance Network (EURGen-Net) capacity survey group (2019) Worsening epidemiological situation of carbapenemase producing Enterobacteriaceae in Europe, assessment by national experts from 37 countries, July 2018. Euro Surveill 24(9):1900123. https://doi.org/10.2807/1560-7917.ES.2019.24.9.1900123
van Beek J, Räisänen K, BroasM Kauranen J, Kähkölä A, Laine J, Mustonen E, Nurkkala T, Puhto T, Sinkkonen J, Torvinen S, Vornanen T, Vuento R, Jalava J, Lyytikäinen O (2019) Tracing local and regional clusters of carbapenemase-producing Klebsiella pneumoniae ST512 with whole genome sequencing, Finland, 2013 to 2018. Euro Surveill 24(38):1800522. https://doi.org/10.2807/1560-7917.ES.2019.24.38.1800522
European Centre for Disease Prevention and Control (2019) Surveillance of antimicrobial resistance in Europe 2018. ECDC, Stockholm. https://doi.org/10.2900/22212
Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, Suppes R, Feinstein D, Zanotti S, Taiberg L, Gurka D, Kumar A, Cheang M (2006) Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 34:1589–1596. https://doi.org/10.1097/01.CCM.0000217961.75225.E9
Acknowledgements
We acknowledge the personnel of HUSLAB Department of Clinical Microbiology for performing the FilmArray BCID2 panel runs.
Funding
Open access funding provided by University of Helsinki including Helsinki University Central Hospital. This study was carried out alongside routine diagnostics.
Author information
Authors and Affiliations
Contributions
Tanja Holma, Jenni Antikainen, and Jari J Martelin conceptualized the study and performed the experiments. Eveliina Tarkka and Tanja Holma collected and analyzed the data as well as supervised the study. Nathlie Friberg, Annika Nevalainen, and Jukka Torvikoski provided clinical expertise for the data analyses and critical comments for the manuscript. Jari J Martelin drafted the manuscript to which all authors amended and agreed to the publication.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The patient samples were collected only from excessive material in the diagnostics. No patients were recruited to the study, and no extra samples were taken from the patients for this purpose. In addition, all samples were processed anonymized for this study. Thus, permission from the ethics committee and informed consents were not required.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: The originally published article was published with errors and missing information in the body text due to lapses during submission process.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Holma, T., Torvikoski, J., Friberg, N. et al. Rapid molecular detection of pathogenic microorganisms and antimicrobial resistance markers in blood cultures: evaluation and utility of the next-generation FilmArray Blood Culture Identification 2 panel. Eur J Clin Microbiol Infect Dis 41, 363–371 (2022). https://doi.org/10.1007/s10096-021-04314-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-021-04314-2