Abstract
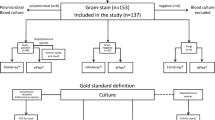
High accuracy of direct from positive blood culture molecular panels is imperative, particularly for the detection of resistance determinants as it allows for antimicrobial optimization prior to conventional susceptibility testing. In this study, we provide extensive data since implementation of the Verigene Gram-positive blood culture panel (BC-GP) in 2013. Within 5 years, 1636 blood culture bottles positive for a Gram-positive organism were tested on the BC-GP panel. The BC-GP panel identified 1520 Gram-positive organisms in 1636 (92.9%) blood cultures tested. For positive blood cultures, we observed 96.4% (806/834) concordance to the species level. Compared with conventional antimicrobial susceptibility testing, the positive percent agreement (PPA) of methicillin-resistant SA (MRSA) (50) and methicillin-resistant SE (MRSE) (365) was 100%. The mecA gene was detected in two methicillin-susceptible Staphylococcus aureus (MSSA) and one methicillin-susceptible S. epidermidis (MSSE) with a negative percent agreement (NPA) of 99.1% (221/223) and 99.2% (120/121), respectively. The PPA and NPA for vancomycin-resistant Enterococcus faecium (VRE) was 100%. The BC-GP panel demonstrated excellent performance and clinicians can confidently de-escalate antimicrobial therapy in the absence of mecA and vanA/B gene.
Similar content being viewed by others
References
Laupland KB (2013) Incidence of bloodstream infection: a review of population-based studies. Clin Microbiol Infect 19(6):492–500
Leibovici L, Shraga I, Drucker M, Konigsberger H, Samra Z, Pitlik SD (1998) The benefit of appropriate empirical antibiotic treatment in patients with bloodstream infection. J Intern Med 244(5):379–386
Spencer DH, Sellenriek P, Burnham CA (2011) Validation and implementation of the GeneXpert MRSA/SA blood culture assay in a pediatric setting. Am J Clin Pathol 136(5):690–694
Rand KH, Delano JP (2014) Direct identification of bacteria in positive blood cultures: comparison of two rapid methods, FilmArray and mass spectrometry. Diagn Microbiol Infect Dis 79(3):293–297
Mestas J, Polanco CM, Felsenstein S, Dien Bard J (2014) Performance of the Verigene Gram-positive blood culture assay for direct detection of Gram-positive organisms and resistance markers in a pediatric hospital. J Clin Microbiol 52(1):283–287
Walker T, Dumadag S, Lee CJ, Lee SH, Bender JM, Cupo Abbott J, She RC (2016) Clinical impact of laboratory implementation of Verigene BC-GN microarray-based assay for detection of gram-negative bacteria in positive blood cultures. J Clin Microbiol 54(7):1789–1796
Roshdy DG, Tran A, LeCroy N, Zeng D, Ou FS, Daniels LM, Weber DJ, Alby K, Miller MB (2015) Impact of a rapid microarray-based assay for identification of positive blood cultures for treatment optimization for patients with streptococcal and enterococcal bacteremia. J Clin Microbiol 53(4):1411–1414
Huang AM, Newton D, Kunapuli A, Gandhi TN, Washer LL, Isip J, Collins CD, Nagel JL (2013) Impact of rapid organism identification via matrix-assisted laser desorption/ionization time-of-flight combined with antimicrobial stewardship team intervention in adult patients with bacteremia and candidemia. Clin Infect Dis 57(9):1237–1245
Samuel LP, Tibbetts RJ, Agotesku A, Fey M, Hensley R, Meier FA (2013) Evaluation of a microarray-based assay for rapid identification of Gram-positive organisms and resistance markers in positive blood cultures. J Clin Microbiol 51(4):1188–1192
Wojewoda CM, Sercia L, Navas M, Tuohy M, Wilson D, Hall GS, Procop GW, Richter SS (2013) Evaluation of the Verigene Gram-positive blood culture nucleic acid test for rapid detection of bacteria and resistance determinants. J Clin Microbiol 51(7):2072–2076
Sullivan KV, Turner NN, Roundtree SS, Young S, Brock-Haag CA, Lacey D, Abuzaid S, Blecker-Shelly DL, Doern CD (2013) Rapid detection of Gram-positive organisms by use of the Verigene Gram-positive blood culture nucleic acid test and the BacT/Alert Pediatric FAN system in a multicenter pediatric evaluation. J Clin Microbiol 51(11):3579–3584
Martinez RM, Bauerle ER, Fang FC, Butler-Wu SM (2014) Evaluation of three rapid diagnostic methods for direct identification of microorganisms in positive blood cultures. J Clin Microbiol 52(7):2521–2529
Siu GK, Chen JH, Ng TK, Lee RA, Fung KS, To SW, Wong BK, Cheung S, Wong IW, Tam MM, Lee SS, Yam WC (2015) Performance evaluation of the Verigene Gram-positive and gram-negative blood culture test for direct identification of bacteria and their resistance determinants from positive blood cultures in Hong Kong. PLoS One 10(10):e0139728
CLSI (2017) Performance standards for antimicrobial Susceptebility testing: twenty-seventh informational supplement M100-S27. CLSI
Buchan BW, Ginocchio CC, Manii R, Cavagnolo R, Pancholi P, Swyers L, Thomson RB Jr, Anderson C, Kaul K, Ledeboer NA (2013) Multiplex identification of gram-positive bacteria and resistance determinants directly from positive blood culture broths: evaluation of an automated microarray-based nucleic acid test. PLoS Med 10(7):e1001478
Kawamura Y, Hou XG, Sultana F, Miura H, Ezaki T (1995) Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int J Syst Bacteriol 45(2):406–408
Petti CA, Woods CW, Reller LB (2005) Streptococcus pneumoniae antigen test using positive blood culture bottles as an alternative method to diagnose pneumococcal bacteremia. J Clin Microbiol 43(5):2510–2512
Bauer KA, West JE, Balada-Llasat JM, Pancholi P, Stevenson KB, Goff DA (2010) An antimicrobial stewardship program's impact with rapid polymerase chain reaction methicillin-resistant Staphylococcus aureus/S. aureus blood culture test in patients with S. aureus bacteremia. Clin Infect Dis 51(9):1074–1080
Sango A, McCarter YS, Johnson D, Ferreira J, Guzman N, Jankowski CA (2013) Stewardship approach for optimizing antimicrobial therapy through use of a rapid microarray assay on blood cultures positive for Enterococcus species. J Clin Microbiol 51(12):4008–4011
Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, Suppes R, Feinstein D, Zanotti S, Taiberg L, Gurka D, Kumar A, Cheang M (2006) Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 34(6):1589–1596
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
This study was not funded by an outside source. J.D.B is has received research funding from Luminex Corporation for other studies not related to this work. All other authors have no reported conflicts of interest. J.M. has collaborated with Luminex Corporation on clinical studies outside the submitted work.
Ethical approval
This study was approved by Children’s Hospital Los Angeles Institutional Review Board (ID: CHLA-18-00071).
Informed consent
Not applicable as per CHLA Institutional Review Board (CHLA-18-0071)
Rights and permissions
About this article
Cite this article
Vareechon, C., Mestas, J., Polanco, C.M. et al. A 5-year study of the performance of the Verigene Gram-positive blood culture panel in a pediatric hospital. Eur J Clin Microbiol Infect Dis 37, 2091–2096 (2018). https://doi.org/10.1007/s10096-018-3343-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-018-3343-2