Abstract
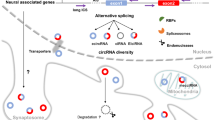
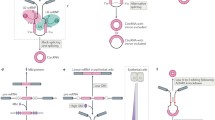
Circular RNAs (circRNAs) provide a new and relatively unexplored class of noncoding RNAs that are predominantly found in mammalian cells. In this review, we present the latest data regarding the structural organization, possible mechanisms of synthesis, and functions of circRNAs. These transcripts were isolated as an RNA fraction that was resistant to RNase R treatment, which selectively destroys the linear forms of RNA molecules. circRNAs are encoded by orthologous genes in different organisms and show tissue- and organ-specific expression. Currently, the biogenesis and functional properties of circRNAs remain unclear; transcripts of this class, however, remain highly promising targets of research. Some of them have been ascribed the function of “molecular sponges” that can absorb microRNAs, RNA-binding proteins, and small nuclear RNAs. circRNAs are often formed from the RNA portions of protein-coding genes in the course of alternative splicing. Some features of the circRNAs of mammals were demonstrated using 11 circRNAs of the human sphingomyelin synthase 1 gene (SGMS1), which were discovered by us in the brain. These circRNAs consist mainly of portions of the multi-exon 5′ untranslated region (5′UTR) of the SGMS1 gene and include one to five exons. The synthesis of circRNAs may be new, previously unknown, function of the multi-exon 5′UTR of genes. This feature is most clearly manifested in the brain, where the level of circRNAs is significantly higher.


Similar content being viewed by others
References
Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO (2013) Cell-type specific features of circular RNA expression. PLoS Genet 9(9):e1003777. doi:10.1371/journal.pgen.1003777
Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO (2012) Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7(2):e30733. doi:10.1371/journal.pone.0030733
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19(2):141–57. doi:10.1261/rna.035667.112
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495(7441):333–8. doi:10.1038/nature11928
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S (2014) CircRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56(1):55–66. doi:10.1016/j.molcel.2014.08.019
Guo JU, Agarwal V, Guo H, Bartel DP (2014) Expanded identification and characterization of mammalian circular RNAs. Genome Biol 15(7):409. doi:10.1186/s13059-014-0409-z
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L (2014) Complementary sequence-mediated exon circularization. Cell 159(1):134–47. doi:10.1016/j.cell.2014.09.001
Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Öhman M, Refojo D, Kadener S, Rajewsky N (2015) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58(5):870–85. doi:10.1016/j.molcel.2015.03.027
Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A (2006) Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res 34(8):e63
Suzuki H, Tsukahara T (2014) A view of pre-RNA splicing from RNase R resistant RNAs. Int J Mol Sci 15(6):9331–42. doi:10.3390/ijms15069331
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL (2013) Circular intronic long noncoding RNAs. Mol Cell 51(6):792–806. doi:10.1016/j.molcel.2013.08.017
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G (2015) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22(3):256–64. doi:10.1038/nsmb.2959
Lasda E, Parker R (2014) Circular RNAs: diversity of form and function. RNA 20(12):1829–42. doi:10.1261/rna.047126.114
Zaphiropoulos PG (1996) Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci U S A 93(13):6536–41
Zaphiropoulos PG (1997) Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol Cell Biol 17(6):2985–93
Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A (2015) Exon circularization requires canonical splice signals. Cell Rep 10(1):103–11. doi:10.1016/j.celrep.2014
Wang Y, Wang Z (2015) Efficient backsplicing produces translatable circular mRNAs. RNA 21(2):172–9. doi:10.1261/rna.048272.114
Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang JL, Yang L, Chen LL (2016) The biogenesis of nascent circular RNAs. Cell Rep 15(3):611–24. doi:10.1016/j.celrep.2016.03.058
Shepard S, McCreary M, Fedorov A (2009) The peculiarities of large intron splicing in animals. PLoS One 4(11):e7853. doi:10.1371/journal.pone.0007853
Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R (1993) Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73(5):1019–30
Cao QP, Gaudette MF, Robinson DH, Crain WR (1995) Expression of the mouse testis-determining gene Sry in male preimplantation embryos. Mol Reprod Dev 40(2):196–204
Wilusz JE, Sharp PA (2013) Molecular biology. A circuitous route to noncoding RNA. Science 340(6131):440–1. doi:10.1126/science.1238522
Jeck WR, Sharpless NE (2014) Detecting and characterizing circular RNAs. Nat Biotechnol 32(5):453–61. doi:10.1038/nbt.2890
Liang D, Wilusz JE (2014) Short intronic repeat sequences facilitate circular RNA production. Genes Dev 28(20):2233–47. doi:10.1101/gad.251926.114
Wilusz JE (2015) Repetitive elements regulate circular RNA biogenesis. Mob Genet Elements 5(3):1–7
Filippenkov IB, Sudarkina OY, Limborska SA, Dergunova LV (2015) Circular RNA of the human sphingomyelin synthase 1 gene: multiple splice variants, evolutionary conservatism and expression in different tissues. RNA Biol 12(9):1030–42. doi:10.1080/15476286.2015.1076611
Vladychenskaya IP, Dergunova LV, Dmitrieva VG, Limborska SA (2004) Human gene MOB: structure specification and aspects of transcriptional activity. Gene 338(2):257–65
Rozhkova AV, Dmitrieva VG, Zhapparova ON, Sudarkina OY, Nadezhdina ES, Limborska SA, Dergunova LV (2011) Human sphingomyelin synthase 1 gene (SMS1): organization, multiple mRNA splice variants and expression in adult tissues. Gene 481(2):65–75. doi:10.1016/j.gene.2011.04.010
Rozhkova AV, Filippenkov IB, Sudarkina OY, Limborska SA, Dergunova LV (2015) Alternative promoters located in SGMS1 gene introns participate in regulation of its expression in human tissues. Mol Biol (Mosk) 49(2):325–33
Pasman Z, Been MD, Garcia-Blanco MA (1996) Exon circularization in mammalian nuclear extracts. RNA 2(6):603–10
Kameyama T, Suzuki H, Mayeda A (2012) Re-splicing of mature mRNA in cancer cells promotes activation of distant weak alternative splice sites. Nucleic Acids Res 40(16):7896–906. doi:10.1093/nar/gks520
Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ (2015) The RNA binding protein quaking regulates formation of circRNAs. Cell 160(6):1125–34. doi:10.1016/j.cell.2015.02.014
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495(7441):384–8. doi:10.1038/nature11993
You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, Wang X, Hou J, Liu H, Sun W, Sambandan S, Chen T, Schuman EM, Chen W (2015) Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 18(4):603–10. doi:10.1038/nn.3975
Herculano-Houzel S (2009) The human brain in numbers: a linearly scaled-up primate brain. Front Hum Neurosci 3:31. doi:10.3389/neuro.09.031.2009
Daniel C, Silberberg G, Behm M, Öhman M (2014) Alu elements shape the primate transcriptome by cis-regulation of RNA editing. Genome Biol 15(2):R28. doi:10.1186/gb-2014-15-2-r28
Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, Lieberman J, Rigoutsos I, Pandolfi PP (2011) Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 147(2):344–57. doi:10.1016/j.cell.2011.09.029
Broderick JA, Zamore PD (2014) Competitive endogenous RNAs cannot alter microRNA function in vivo. Mol Cell 54(5):711–3. doi:10.1016/j.molcel.2014.05.023
Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M (2014) Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell 54(5):766–76. doi:10.1016/j.molcel.2014.03.045
Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J (2011) miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 30(21):4414–22. doi:10.1038/emboj.2011.359
Hansen TB, Kjems J, Damgaard CK (2013) Circular RNA and miR-7 in Cancer. Cancer Res 73(18):5609–12. doi:10.1158/0008-5472.CAN-13-1568
Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S, Purow B (2008) microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res 68(10):3566–72. doi:10.1158/0008-5472
Jiang L, Liu X, Chen Z, Jin Y, Heidbreder CE, Kolokythas A, Wang A, Dai Y, Zhou X (2010) microRNA-7 targets IGF1R (insulin-like growth factor 1 receptor) in tongue squamous cell carcinoma cells. Biochem J 432(1):199–205. doi:10.1042/BJ20100859
Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM (2009) Repression of alphasynuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci U S A 106(31):13052–7. doi:10.1073/pnas.0906277106
Huang C, Shan G (2015) What happens at or after transcription: insights into circRNA biogenesis and function. Transcription 6(4):61–4. doi:10.1080/21541264.2015.1071301
Veno MT, Hansen TB, Veno ST, Clausen BH, Grebing M, Finsen B, Holm IE, Kjems J (2015) Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol 16:245. doi:10.1186/s13059-015-0801-3
Somers J, Pöyry T, Willis AE (2013) A perspective on mammalian upstream open reading frame function. Int J Biochem Cell Biol 45(8):1690–700. doi:10.1016/j.biocel.2013.04.020
Sudarkina OY, Filippenkov IB, Brodsky IB, Limborska SA, Dergunova LV (2015) Comparative analysis of sphingomyelin synthase 1 gene expression at the transcriptional and translational levels in human tissues. Mol Cell Biochem 406(1-2):91–9. doi:10.1007/s11010-015-2427-x
Acknowledgments
We thank the Russian Foundation for Basic Research for support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Funding
This work was supported by a grant from the Molecular and Cellular Biology Program of the Russian Academy of Sciences and by grant from the Russian Foundation for Basic Research (grant number 16-04-00488; grant number 16-34-00653).
Rights and permissions
About this article
Cite this article
Filippenkov, I.B., Kalinichenko, E.O., Limborska, S.A. et al. Circular RNAs—one of the enigmas of the brain. Neurogenetics 18, 1–6 (2017). https://doi.org/10.1007/s10048-016-0490-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-016-0490-4