Abstract
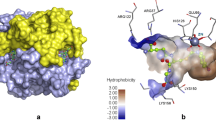
Biguanide derivatives exhibit a wide variety of therapeutic applications, including anti-cancer effects. Metformin is an effective anti-cancer agent against breast cancer, lung cancer, and prostate cancer. In the crystal structure (PDB ID: 5G5J), it was found that metformin is found in the active site of CYP3A4, and the associated anti-cancer effect was explored. Taking clues from this work, pharmacoinformatics research has been carried out on a series of known and virtual biguanide, guanylthiourea (GTU), and nitreone derivatives. This exercise led to the identification of more than 100 species that exhibit greater binding affinity toward CYP3A4 in comparison to that of metformin. Selected six molecules were subjected to molecular dynamics simulations, and the results are presented in this work.










Similar content being viewed by others
Data availability
Electronic Supporting Information (ESI) was uploaded. Additional supporting information may be found in the online version of this article.
References
Kathuria D, Raul AD, Wanjari P, Bharatam PV (2021) Biguanides: species with versatile therapeutic applications. Eur J Med Chem 219:113378. https://doi.org/10.1016/j.ejmech.2021.113378
Kathuria D, Bankar AA, Bharatam PV (2018) “What’s in a structure?” The story of biguanides. J Mol Struct 1152:61–78. https://doi.org/10.1016/j.molstruc.2017.08.100
Pollak M (2010) Metformin and other biguanides in oncology: advancing the research agendametformin and other biguanides in oncology. Cancer Prev Res (Phila) 3:1060–1065. https://doi.org/10.1158/1940-6207.CAPR-10-0175
Bailey CJ (1992) Biguanides and NIDDM. Diabetes Care 15:755–772. https://doi.org/10.2337/diacare.15.6.755-72
Slotta K, Über TR (1929) Biguanide, II.: Die blutzucker-senkende Wirkung der Biguanide. Berichte der deutschen chemischen Gesellschaft (A and B Series) 62:1398–1405. https://doi.org/10.1002/cber.19290620605
Curd F, Davey D, Rose F (1945) Studies on synthetic antimalarial drugs: X.—some biguanide derivatives as new types of antimalarial substances with both therapeutic and causal prophylactic activity. Ann Trop Med Parasitol 39:208–216. https://doi.org/10.1080/00034983.1945.11685237
Ungar G, Freedman L, Shapiro SL (1957) Pharmacological studies of a new oral hypoglycemic drug. Proc Soc Exp Biol Med 95:190–192. https://doi.org/10.3181/00379727-95-23163
Mehnert H, Seitz W (1958) Weitere Ergebnisse der diabetesbehandlung mit blutzuckersenkendenbiguaniden. Munch Med Wschr 100:1849–1851
Barnes GP, Carter HG, Gross A, Bhaskar SN, Schildt NN, Bush AG (1972) Dental plaque reduction with an antibacterial mouth rinse. Part I. Oral Surg Oral Med Oral Pathol 34:553–558. https://doi.org/10.1016/0030-4220(72)90337-4
Pinelli A, Trivulzio S, Pojaga G, Rossoni G (1996) Effects of 2-guanidine-4-methylquinazoline on gastric acid secretion in rats. Pharmacol Res 34:225–230. https://doi.org/10.1006/phrs.1996.0092
García Rubiño ME, Carrillo E, Ruiz Alcalá G, Domínguez-Martín A, Marchal A, Boulaiz J, H. (2019) Phenformin as an anticancer agent: challenges and prospects. Int J Mol Sci 20:3316. https://doi.org/10.3390/ijms20133316
Sharma P, Kumar S (2018) Metformin inhibits human breast cancer cell growth by promoting apoptosis via a ROS-independent pathway involving mitochondrial dysfunction: pivotal role of superoxide dismutase (SOD). Cell Oncol 41:637–650. https://doi.org/10.1007/s13402-018-0398-0
Henderson D, Frieson D, Zuber J, Solomon SS (2017) Metformin has positive therapeutic effects in colon cancer and lung cancer. Am J Med Sci 354:246–251. https://doi.org/10.1016/j.amjms.2017.05.006
Li L, Wang L, Li J, Fan Z, Yang L, Zhang Z et al (2018) Metformin-induced reduction of CD39 and CD73 blocks myeloid-derived suppressor cell activity in patients with ovarian cancer. Cancer Res 78:1779–1791. https://doi.org/10.1158/0008-5472.CAN-17-2460
Wahdan-Alaswad RS, Cochrane DR, Spoelstra NS, Howe EN, Edgerton SM, Anderson SM et al (2014) Metformin-induced killing of triple-negative breast cancer cells is mediated by reduction in fatty acid synthase via miRNA-193b. Horm Cancer 5:374–389. https://doi.org/10.1007/s12672-014-0188-8
Sharma A, Bandyopadhayaya S, Chowdhury K, Sharma T, Maheshwari R, Das A et al (2019) Metformin exhibited anticancer activity by lowering cellular cholesterol content in breast cancer cells. PLoS One. 14:e0209435. https://doi.org/10.1371/journal.pone.0209435
Zhou S, Xu L, Cao M, Wang Z, Xiao D, Xu S et al (2019) Anticancer properties of novel pyrazole-containing biguanide derivatives with activating the adenosine monophosphate- activated protein kinase signaling pathway. Arch Pharm 352:1900075. https://doi.org/10.1002/ardp.201900075
Mitra R, Guo Z, Milani M, Mesaros C, Rodriguez M, Nguyen J et al (2011) CYP3A4 mediates growth of estrogen receptor-positive breast cancer cells in part by inducing nuclear translocation of phospho-Stat3 through biosynthesis of (±)-14, 15-epoxyeicosatrienoic acid (EET). J Biol Che 286:17543–17559. https://doi.org/10.1074/jbc.M110.198515
Guo Z, Johnson V, Barrera J, Porras M, Hinojosa D, Hernández I et al (2018) Targeting cytochrome P450-dependent cancer cell mitochondria: cancer associated CYPs and where to find them. Cancer and Metastasis Revi 37:409–423
Guo Z, Sevrioukova IF, Denisov IG, Zhang X, Chiu TL, Thomas DG et al (2017) Heme binding biguanides target cytochrome P450-dependent cancer cell mitochondria. Cell Chem Bio 24(1259–75):e6. https://doi.org/10.1016/j.chembiol.2017.08.009
Bharatam PV, Patel DS, Iqbal P (2005) Pharmacophoric features of biguanide derivatives: an electronic and structural analysis. J Med Chem 48:7615–7622. https://doi.org/10.1021/jm050602z
Patel DS, Bharatam PV (2009) Novel⊕N (← L) 2 species with two lone pairs on nitrogen: systems isoelectronic to carbodicarbenes. Chem Commun 1064-6. https://doi.org/10.1039/B816595E
Patel DS, Bharatam PV (2011) Divalent N (I) compounds with two lone pairs on nitrogen. J Phy Chem A 115:7645–7655. https://doi.org/10.1021/jp111017u
Bharatam PV, Arfeen M, Patel N, Jain P, Bhatia S, Chakraborti AK et al (2016) Design, synthesis, and structural analysis of divalent NI compounds and identification of a new electron-donating ligand. Chem Euro J 22:1088–1096. https://doi.org/10.1002/chem.201503618
Patel N, Arfeen M, Singh T, Bhagat S, Sakhare A, Bharatam PV (2020) Divalent NI compounds: identifying new carbocyclic carbenes to design nitreones using quantum chemical methods. J Comput Chem 41:2624–2633. https://doi.org/10.1002/jcc.26417
Bhatia S, Malkhede YJ, Bharatam PV (2013) Existence of dynamic tautomerism and divalent N (I) character in N-(pyridin-2-yl) thiazol-2-amine. J Comput Chem 34:1577–1588. https://doi.org/10.1002/jcc.23293
Bhagat S, Arfeen M, Das G, Patel N, Bharatam PV (2019) Electronic and ligating properties of carbocyclic carbenes: a theoretical investigation. J Comput Chem 40:726–733. https://doi.org/10.1002/jcc.25756
Wanjari PJ, Singh T, Sofi F, Bharatam PV (2021) Quantum chemical study in exploring the role of donor→ acceptor interactions in 1, 3-bis carbene-stabilized guanidinium cations. J Mole Model 27:1–10. https://doi.org/10.1007/s00894-021-04707-2
Dar MO, Dubey G, Singh T, Bharatam PV (2022) N-heterocyclic carbene ligated oximes: exploring the electronic structure and properties. Int J Quantum Chem 122:e26907. https://doi.org/10.1002/qua.26907
Singh S, Wanjari PJ, Bhatia S, Sonwane VC, Chakraborti AK, Bharatam PV (2015) Design, synthesis, biological evaluation and toxicity studies of N, N-disubstituted biguanides as quorum sensing inhibitors. Med Chem Res 24:1974–1987. https://doi.org/10.1007/s00044-014-1255-y
Singh T, George A, Parameswaran P, Bharatam PV (2019) Enols, diamino enols, and breslow intermediates: a comparative quantum chemical analysis. Eur J Org Chem 2019:2481–2489. https://doi.org/10.1002/ejoc.201801817
Singh T, Sahoo SC, Bharatam PV (2021) Compound with possible N→ N coordination bond: synthesis, crystal structure and electronic structure analysis. Tet Lett 77:153246. https://doi.org/10.1016/j.tetlet.2021.153246
Arfeen M, Patel DS, Abbat S, Taxak N, Bharatam PV (2014) Importance of cytochromes in cyclization reactions: quantum chemical study on a model reaction of proguanil to cycloguanil. J Comput Chem 35:2047–2055. https://doi.org/10.1002/jcc.23719
Dubey G, Mahawa N, Singh T, Saha N, Sahoo SC, Bharatam PV (2022) Thiazetidin-2- ylidenes as four membered N-heterocyclic carbenes: theoretical studies and the generation of complexes with N+ center. Phys Chem Chem Phys 24:629–633. https://doi.org/10.1039/D1CP04732A
Patel DS, Bharatam PV (2011) To bend or not to bend! The dilemma of allenes. J Org Chem 76:2558–2567. https://doi.org/10.1021/jo102432a
Singh T, Bharatam PV (2019) Donor→ acceptor coordination interactions in 1, 3-bis (NHC) triazenyl cations: an electronic structure analysis. J Comput Chem 40:2207–2215. https://doi.org/10.1002/jcc.25872
Bhagat S, Arfeen M, Adane L, Singh S, Singh PP, Chakraborti AK et al (2017) Guanylthiourea derivatives as potential antimalarial agents: synthesis, in vivo and molecular modelling studies. Eur J Med Chem 135:339–348. https://doi.org/10.1016/j.ejmech.2017.04.022
Adane L, Bhagat S, Arfeen M, Bhatia S, Sirawaraporn R, Sirawaraporn W et al (2014) Design and synthesis of guanylthiourea derivatives as potential inhibitors of Plasmodium falciparum dihydrofolate reductase enzyme. Bioorg Med Chem Lett 24:613–617. https://doi.org/10.1016/j.bmcl.2013.12.009
Adane L, Bharatam PV (2011) Computer-aided molecular design of 1H-imidazole-2, 4- diamine derivatives as potential inhibitors of Plasmodium falciparum DHFR enzyme. J Mol Model 17:657–667. https://doi.org/10.1007/s00894-010-0756-y
Bhagat S, Arfeen M, Das G, Ramkumar M, Khan SI, Tekwani BL et al (2019) Design, synthesis and biological evaluation of 4-aminoquinoline-guanylthiourea derivatives as antimalarial agents. Bioor Chemistry. 91:103094. https://doi.org/10.1016/j.bioorg.2019.103094
Mehdi A, Adane L, Patel DS, Bharatam PV (2010) Electronic structure and reactivity of guanylthiourea: a quantum chemical study. J Comput Chem 31:1259–1267. https://doi.org/10.1002/jcc.21412
Patel N, Arfeen M, Sood R, Khullar S, Chakraborti AK, Mandal SK et al (2018) Can remote N-heterocyclic carbenes coordinate with main group elements? Synthesis, structure, and quantum chemical analysis of N+centered complexes. Chem Eur J 24:6418–6425. https://doi.org/10.1002/chem.201705999
Patel N, Sood R (2018) Bharatam PV NL2+ systems as new-generation phase-transfer catalysts. Chem Rev 118:8770–8785. https://doi.org/10.1021/acs.chemrev.8b00169
Dubey G, Awari S, Singh T, Sahoo SC, Bharatam PV (2021) Mesoionic and N-heterocyclic carbenes coordinated N+ center: experimental and computational analysis. ChemPlusChem 86:1416–1420. https://doi.org/10.1002/cplu.202100281
Bhatia S, Bagul C, Kasetti Y, Patel DS, Bharatam PV (2012) Divalent N (I) character in 2- (thiazol-2-yl) guanidine: an electronic structure analysis. J Phy Chem A 116:9071–9079. https://doi.org/10.1021/jp304789u
Patel N, Falke B, Bharatam PV (2018) C→ N coordination bonds in (CCC)→ N+←(L) complexes. Theor Chem Acc 137:1–10. https://doi.org/10.1007/s00214-018-2208-1
Bhatia S, Bharatam PV (2014) Possibility of the existence of donor–acceptor interactions in bis (azole) amines: an electronic structure analysis. J Org Chem 79:4852–4862. https://doi.org/10.1021/jo402862r
Kathuria D, Arfeen M, Bankar AA, Bharatam, (2016) PV Carbene→ N+ coordination bonds in drugs: a quantum chemical study. J Chem Sci 128:1607–1614. https://doi.org/10.1007/s12039-016-1173-2
Lokhande K, Venkateswara NNK, S, Pawar S. (2022) Biflavonoids from Rhus succedanea as probable natural inhibitors against SARS-CoV-2: a molecular docking and molecular dynamics approach. J Biomol Struct Dyn 40:4376–4388. https://doi.org/10.1080/07391102.2020.1858165
Didziapetris R, Dapkunas J, Sazonovas A, Japertas P (2010) Trainable structure–activity relationship model for virtual screening of CYP3A4 inhibition. J Comput Aided Mol Des 24:891–906. https://doi.org/10.1007/s10822-010-9381-1
Crivori P, Zamora I, Speed B, Orrenius C, Poggesi I (2004) Model based on GRID-derived descriptors for estimating CYP3A4 enzyme stability of potential drug candidates. J Comput Aided Mol Des 18:155–166. https://doi.org/10.1023/B:JCAM.0000035184.11906.c2
Kriegl JM, Arnhold T, Beck B, Fox T (2005) A support vector machine approach to classify human cytochrome P450 3A4 inhibitors. J Comput Aided Mol Des 19:189–201. https://doi.org/10.1007/s10822-005-3785-3
Cherkaoui-Malki M, Meyer K, Cao WQ, Latruffe N, Yeldandi AV, Rao MS et al (2001) Identification of novel peroxisome proliferator-activated receptor α (PPARα) target genes in mouse liver using cDNA microarray analysis. Gene Expr J Liver Res 9:291–304. https://doi.org/10.3727/000000001783992533
Shaikh F, Bhakat S, Thakur A, Radadia A, Soliman ES, M, Shah A, (2015) Identification of novel GSK1070916 analogs as potential aurora b inhibitors: insights from molecular dynamics and MM/GBSA based rescoring. Lett Drug Des & Discov 12:2–13
Patel DC, Hausman KR, Arba M, Tran A, Lakernick PM, Wu C (2022) Novel inhibitors to ADP ribose phosphatase of SARS-CoV-2 identified by structure-based high throughput virtual screening and molecular dynamics simulations. Comput Biol Med 140:105084. https://doi.org/10.1016/j.compbiomed.2021.105084
Lokhande K, Venkateswara NNK, S, Pawar S, (2022) Biflavonoids from Rhus succedanea as probable natural inhibitors against SARS-CoV-2: a molecular docking and molecular dynamics approach. J Biomol Struct Dyn 40:4376–4388. https://doi.org/10.1080/07391102.2020.1858165
Rajagopal K, Byran G, Jupudi S, Vadivelan R (2020) Activity of phytochemical constituents of black pepper, ginger, and garlic against coronavirus (COVID-19): an in silico approach. Int J Health Allied Sci 9:43–50. https://doi.org/10.4103/ijhas.IJHAS_55_20
Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA et al (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein−ligand complexes. J Med Chem 49:6177–6196. https://doi.org/10.1021/jm051256o
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT et al (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of dockingm accuracy. J Med Chem 47:1739–1749. https://doi.org/10.1021/jm0306430
Benet LZ, Hosey CM, Ursu O, Oprea TI (2016) BDDCS, the rule of 5 and drugability. Adv Drug Deliv Rev 101:89–98. https://doi.org/10.1016/j.addr.2016.05.007
Deng N, Forli S, He P, Perryman A, Wickstrom L, Vijayan R et al (2015) Distinguishing binders from false positives by free energy calculations: fragment screening against the flap site of HIV protease. J Phys Chem B 119:976–988. https://doi.org/10.1021/jp506376z
Rego CM, Francisco AF, Boeno CN, Paloschi MV, Lopes JA, Silva MD et al (2022) Inflammasome NLRP3 activation induced by convulxin, a C-type lectin-like isolated from Crotalus durissus terrificus snake venom. Sci Rep 12:4706. https://doi.org/10.1038/s41598-022-08735-7
Lima AdM, Siqueira AS, Möller MLS, Souza RCd, Cruz JN, Lima ARJ, et al (2022) In silico improvement of the cyanobacterial lectin microvirin and mannose interaction. J Biomol Struct Dyn, 40 1064-73. https://doi.org/10.1080/07391102.2020.1821782
Almeida VM, Dias ÊR, Souza BC, Cruz JN, Santos CB, Leite FH et al (2022) Methoxylated flavonols from Vellozia dasypus Seub ethyl acetate active myeloperoxidase extract: in vitro and in silico assays. J Biomol Struct Dyn 40:7574–7583. https://doi.org/10.1080/07391102.2021.1900916
Muzammil S, Neves Cruz J, Mumtaz R, Rasul I, Hayat S, Khan MA et al (2023) Effects of drying temperature and solvents on in vitro diabetic wound healing potential of moringa oleifera leaf extracts. Molecules 28:710. https://doi.org/10.3390/molecules28020710
Funding
Department of Biotechnology, Government of India for financial support (grant number: BT/PR140164); SERB, Department of Science and Technology, Government of India for funding (PDF/2020/002825).
Author information
Authors and Affiliations
Contributions
Pravin J. Wanjari: revised the manuscript critically and wrote the manuscript, Asutosh Rath: execution of idea, Rohit Y. Sathe: revised the manuscript critically, and Prasad V. Bharatam: generation of idea, analysis, and revised it critically.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wanjari, P.J., Rath, A., Sathe, R.Y. et al. Identification of CYP3A4 inhibitors as potential anti-cancer agents using pharmacoinformatics approach. J Mol Model 29, 156 (2023). https://doi.org/10.1007/s00894-023-05538-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-023-05538-z