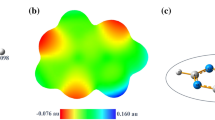
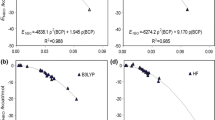
Abstract
The effect of vicinal molecular groups on the intrinsic acidity of a central guanine residue in short single-stranded DNA models and the potentials exerted by the backbone and the nucleobases on the leaving proton were determined by the fragment molecular orbital (FMO) method, in terms of quantum descriptors (QDs) and pair interaction interfragment decomposition analysis (PIEDA). The acidity of the central guanine moiety decreased with increasing oligonucleotide length, in response to changes by less than 1 eV in the ionization potential, global softness, electrophilicity index, and electronegativity descriptors. The differences in these descriptors were majorly interpreted in terms of the electrostatic influence of the negative charges residing on the backbone of the molecule. Additionally, this electric-field effect was determined explicitly for the displacement of the test hydronium ion to a distance of 250 Å from its original position, resulting in good agreement with calculations of the variation in Gibbs free energies, obtained from physical experiments conducted on the identical oligonucleotide sequences. The reported results are useful for biophysical applications of deoxyriboligonucleotides containing guanine residues in order to induce local negative charges at specific positions in the DNA chain.






Similar content being viewed by others
Data Availability
All raw data are available from the authors upon request.
References
Chatterjee S, Pathmasiri W, Plashkevych O, Honcharenko D, Varghese OP, Maiti M, Chattopadhyaya J (2006) The chemical nature of the 2′-substituent in the pentose-sugar dictates the pseudoaromatic character of the nucleobase (pKa) in DNA/RNA. Org Biomol Chem 4:1675–1686
Acharya S, Barman J, Cheruku P, Chatterjee S, Acharya P, Isaksson J, Chattopadhyaya J (2004) Significant pKa perturbation of nucleobases is an intrinsic property of the sequence context in DNA and RNA. J Am Chem Soc 126:8674–8681
Galindo-Murillo R, Bergonzo C, Cheatham III TE (2014) Molecular modeling of nucleic acid structure: energy and sampling. Curr Protoc Nucleic Acid Chem 56:7.10.1. https://doi.org/10.1002/0471142700.nc0708s04
Šponer J, BanአP, Jurečka P, Zgarbová M, Kührová P, Havrila M, Krepl M, Stadlbauer P, Otyepka M (2014) Molecular dynamics simulations of nucleic acids. From tetranucleotides to the ribosome. J Phys Chem Lett 5:1771–1782
Šponer J, Shukla MK, Wang J., Leszczynski J (2017) Computational modeling of DNA and RNA fragments. In: Leszczynski J, Kaczmarek-Kedziera A, Puzyn T, G Papadopoulos M, Reis H, K. Shukla M (eds) Handbook of Computational Chemistry. Springer, Cham. p 1803–1826. https://doi.org/10.1007/978-3-319-27282-5_35
Witts RN, Hopson EC, Koballa DE, Van Boening TA, Hopkins NH, Patterson EV, Nagan MC (2013) Backbone−base interactions critical to quantum stabilization of transfer RNA Anticodon Structure. J Phys Chem 117:7489−7497.
Koch T, Shim I, Lindow M, Ørum H, Bohr HG (2014) Quantum mechanical studies of DNA and LNA. Nucleic Acid Ther 24:139–148
Bravaya KB, Epifanovsky E, Krylov AI (2012) Four bases score a run: ab initio calculations quantify a cooperative effect of H-bonding and π-stacking on the ionization energy of adenine in the AATT tetramer. J Phys Chem Lett 3:2726–2732
Tonzani S, Schatz GC (2008) Electronic excitations and spectra in single-stranded DNA. J Am Chem Soc 130:7607–7612
Kurisaki I, Fukuzawa K, Nakano T, Mochizuki Y, Watanabe H, Tanaka S (2010) Fragment molecular orbital (FMO) study on stabilization mechanism of neuro-oncological ventral antigen (NOVA)-RNA complex system. J Mol Struct 962:45–55
Fukuzawa K, Komeiji Y, Mochizuki Y, Kato A, Nakano T, Tanaka S (2006) Intra- and intermolecular interactions between cyclic-AMP receptor protein and DNA: ab initio fragment molecular orbital study. J Comput Chem 27:948–960
Kurisaki I, Fukuzawa K, Komeiji Y, Mochizuki Y, Nakano T, Imada J, Chmielewski A, Rothstein SM, Watanabe H, Tanaka S (2007) Visualization analysis of inter-fragment interaction energies of CRP–cAMP–DNA complex based on the fragment molecular orbital method. Biophys Chem 130:1–9
Iwata T, Fukuzawa K, Nakajima K, Aida-Hyugaji S, Mochizuki Y, Watanabe H, Tanaka S (2008) Theoretical analysis of binding specificity of influenza viral hemagglutinin to avian and human receptors based on the fragment molecular orbital method. Comput Biol Chem 32:198–211
Mazanetz MP, Ichihara O, Law RJ, Whittaker M (2011) Prediction of cyclin-dependent kinase 2 inhibitor potency using the fragment molecular orbital method. J Cheminf 3:2
Scharner J, Aznarez I (2020) Clinical Applications of Single-Stranded Oligonucleotides: Current Landscape of Approved and In-Development Therapeutics. Mol Ther 29:540–554
Bathe M, Rothemund PWK (2017) DNA nanotechnology: a foundation for programmable nanoscale materials. MRS Bulletin 42:882–888
Gutmanas A, Alhroub Y, Battle GM, Berrisford JM, Bochet E, Conroy MJ, Dana JM, Fernández Montecelo MA, van Ginkel G, Gore SP, Haslam P, Hatherley R, Hendrickx PMS, Hirshberg M, Lagerstedt I, Mir S, Mukhopadhyay A, Oldfield TJ, Patwardhan A et al (2014) PDBe: Protein data bank in Europe. Nucleic Acids Res 42:D285
van Dijk M, Bonvin AM (2009) 3D-DART: a DNA structure modelling server. Nucleic Acids Res. 37(suppl 2):W235–W239
El Hassan MA, Calladine CR (1997) Conformational characteristics of DNA: empirical classifications and a hypothesis for the conformational behaviour of dinucleotide steps. Philos T Roy Soc A 335:43–100
Li H, Fedorov DG, Nagata T, Kitaura K, Jensen JH, Gordon MS (2010) Energy gradients in combined fragment molecular orbital and polarizable continuum model (FMO/PCM) calculation. J Comput Chem 31:778–790
Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH, Koseki S, Matsunaga N, Nguyen KA, Su S, Windus TL, Dupuis M, Montgomery Jr JA (1993) General atomic and molecular electronic structure system. J Comput Chem 14:1347–1363
Suenaga M (2005) Facio: new computational chemistry environment for PC GAMESS. J Comput Chem 4:25–32
Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/
Kitaura K, Ikeo E, Asada T, Nakano T, Uebayasi M (1999) Fragment molecular orbital method: an approximate computational method for large molecules. Chem Phys Lett 313:701–706
Nakano T, Kaminuma T, Sato T, Fukuzawa K, Akiyama Y, Uebayasi M, Kitaura K (2002) Fragment molecular orbital method: use of approximate electrostatic potential. Chem Phys Lett 351:475–480
Fedorov DG, Kitaura K (2007) Pair interaction energy decomposition analysis. J Comput Chem 28:222–237
Tomasi J, Mennucci B, Cammi R (2005) Quantum mechanical continuum solvation models. Chem Rev 105:2999–3093
Fedorov DG, Kitaura K, Li H, Jensen JH, Gordon MS (2006) The polarizable continuum model (PCM) interfaced with the fragment molecular orbital method (FMO). J Comput Chem 27:976–985
Fedorov DG, Kitaura K (2012) Energy decomposition analysis in solution based on the fragment molecular orbital method. J Phys Chem A 116:704–719
Rajak SK, Islam N, Ghosh DC (2011) Modeling of the chemico-physical process of protonation of molecules entailing some quantum chemical descriptors. J Quantum Inf. Sci 1:87–95
Origin (Pro), Version Number (2020b) OriginLab Corporation. Northampton, MA, USA
Koopmans T (1934) Über die Zuordnung von Wellenfunktionen und Eigenwerten zu den einzelnen Elektronen eines Atoms. Physica 1:104–113
Fukui K (1982) Role of frontier orbitals in chemical reactions. Science 218:747–754
Parr RG, Szentpaly LV, Liu S (1999) Electrophilicity index. J Am Chem Soc 121:1922–1924
González-Olvera JC, Martínez-Reyes J, González-Jasso E, Pless RC (2015) Determination of pKa values for deprotonable nucleobases in model oligonucleotides. Biophys Chem 206:58–65
Isaksson J, Acharya S, Barman J, Cheruku P, Chattopadhyaya J (2004) Single-stranded adenine-rich DNA and RNA retain structural characteristics of their respective double-stranded conformations and show directional differences in stacking pattern. Biochemistry 43:15996–16010
Capobianco A, Caruso T, Peluso A (2015) Hole delocalization over adenine tracts in single stranded DNA oligonucleotides. Phys Chem Chem Phys 17:4750–4756
Chakraborty D, Hori N, Thirumalai D (2018) Sequence-dependent three interaction site model for single- and double-stranded DNA. J Chem Theory Comput 14:3763–3779
Capobianco A, Velardo A, Peluso A (2018) Single stranded DNA oligonucleotides retain rise coordinates characteristic of double helices. J Phys Chem B 122:7978–7989
Plumridge A, Meisburger SP, Andresen K, Pollack L (2017) The impact of base stacking on the conformations and electrostatics of single-stranded DNA. Nucleic Acids Res 45:3932–3943
Heck A, Woiczikowski PB, Kubař T, Welke K, Niehaus T, Giese B, Skourtis S, Elstner M, Steinbrecher TB (2014) Fragment orbital based description of charge transfer in peptides including backbone orbitals. J Phys Chem B 118:4261–4272
Fujita T, Mochizuki Y (2018) Development of the fragment molecular orbital method for calculating nonlocal excitations in large molecular systems. J Phys Chem A 122:3886–3898
Chakraborty K, Mantha S, Bandyopadhyaya S (2013) Molecular dynamics simulation of a single-stranded DNA with heterogeneous distribution of nucleobases in aqueous medium. J Chem Phys 139:075103
Chakraborty K, Bandyopadhyay S (2014) Correlated dynamical crossovers of the hydration layer of a single-stranded DNA oligomer. J Phys Chem B 118:413–422
Šponer J, Mládek A, Šponer JE, Svozil D, Zgarbová M, BanአP, Jurečka P, Otyepka M (2014) The DNA and RNA sugar–phosphate backbone emerges as the key player. An overview of quantum-chemical, structural biology and simulation studies. Phys Chem Chem Phys 14:15257–15277
Code availability
All the codes used in the calculations are available from the authors upon request.
Funding
JCGO received support from a Graduate student stipend from CONACYT. The stipend number is 37678. This work is part of CONACYT Basic Science Project No. 61322.
Author information
Authors and Affiliations
Contributions
All authors designed the project. JCGO and RCP performed the calculations. All authors contribute to writing of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 84 kb)
Rights and permissions
About this article
Cite this article
González-Olvera, J.C., Zamorano-Carrillo, A., Arreola-Jardón, G. et al. Residue interactions affecting the deprotonation of internal guanine moieties in oligodeoxyribonucleotides, calculated by FMO methods. J Mol Model 28, 43 (2022). https://doi.org/10.1007/s00894-022-05033-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-022-05033-x