Abstract
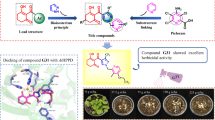
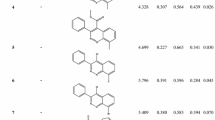
Phalaris minor is a major weed of wheat crop which has evolved resistance against herbicides. Isoproturon is the most accepted herbicide developed resistance in 1992. Later, introduced herbicides also developed resistance and cross-resistance to their respective binding sites. Isoproturon binds at the QB binding site of the D1 protein of photosystem-II (PS-II), which blocks the electron transfer in photosynthesis. In this work, we have carried out a series of computational studies to prioritize the promising herbicides against D1 protein of P. minor. Through the computational studies, twenty-four lead molecules are prioritized which have shown a higher binding affinity and inhibition constant than the reference ligand molecule. The binding and conformational stability of docked complexes was evaluated by molecular dynamics simulations and binding free energy calculations i.e., MM/PBSA. A list of amino acids such as Ala225, Ser226, Phe227, and Asn229 present in the binding site of protein is obtained to be playing an important role in the stability of the protein-lead complex via hydrogen bond and π-π interactions. Binding free energy calculation revealed that the selected lead molecule binding is energetically favorable and driven by electrostatic interactions. Among 24 leads, computational results have uncovered eight promising compounds as potential herbicides which have shown comparable physiochemical profile, better docking scores, system stability, H-bond occupancy, and binding free energy than terbutryn, a reference molecule. These prioritized molecules were custom synthesized and evaluated for their herbicidal activity and specificity through whole plant assay under laboratory-controlled conditions. The lead molecule ELC5 (6-ethoxy-4-N-(2-morpholin-4-ylethyl)-2-N-propan-2-yl-1,3,5-triazine-2,4-diamine) has shown comparable activity to the reference herbicide(isoproturon) against P. minor.
Graphical abstract















Similar content being viewed by others
Availability of data and material
Supplementary information is available for this paper.
Code availability
Not applicable.
7. References
Chhokar RS, Sharma RK (2008) Multiple herbicide resistant in little seed canary grass (Phalaris minor): a threat production in India. Weed Biol Manag 8:112–123
Chhokar RS, Sharma RK, Sharma I (2012) Weed management strategies in wheat-a review. J Wheat Res 4:1–21
Yadav A, Malik RK (2005) Botanical characteristics, biology and distribution in herbicide resistant Phalaris minor in wheat - a sustainability issue. Hisar, India Department of Agronomy and Directorate of Extension Education, CCS Haryana Agricultural University, 5–15
Yadav A, Malik RK (2012) Alternate herbicides against isoproturon resistant Phalaris minor in herbicide resistant Phalaris minor in wheat- a sustainability issue. Hisar, India, Department of Agronomy and Directorate of Extension Education CCS Haryana Agricultural University, 89–98
Singh DV, Gaur AK, Mishra DP (2004) Biochemical and molecular mechanisms of resistance against isoproturon in Phalaris minor: variations in protein and RAPD profiles of isoproturon resistant and sensitive Phalaris minor biotypes. Indian J Weed Sci 36:256–259
Tripathi MK, Yadav MK, Gaur AK, Mishra DP (2005) PCR based isolation of psbA (herbicide binding protein encoding) gene using chloroplast and genomic DNA from Phalaris minor biotype(s). Physiol Mol Biol Plants 11:161–163
Marder JΒ, Chapman DJ, Telfer A, Nixon ΡJ, Barber J (1987) Identification of psbA and psbD gene products, Dl and D2, as reaction centre proteins of photosystem 2. Plant Mol Biol 9:325–333
Horovitz A, Ohad N, Hirschberg J (1989) Predicted effects on herbicide binding of amino acid substitutions in the D1 protein of photosystem II. FEBS Lett 234:161–164
Hirschberg J, Bleeckes A, Kyle DJ, McIntosh L, Arntzen CJ (1984) The molecular basis of triazine resistance in higher plant chloroplasts. Natur Forch 39:412–419
Hirschberg J, Yehuda AB, Pecker I, Ohad N (1987) Mutations resistant to photosystem II herbicides. NATO ASI series. Series A: Life Sciences 140:357–66
Rani P, Kumari J, Agarwal S, Singh DV (2019) Binding mode of aryloxyphenoxypropionate (FOP) and cyclohexanedione (DIM) groups of herbicides at the carboxyl transferase (CT) domain of Acetyl-CoA carboxylase of Phalaris minor. Netw Model Anal Health Inform Bioinforma 8:1–14
Singh DV, Adeppa K, Misra K (2011) Mechanism of isoproturon resistance in Phalaris minor: in silico design, synthesis and testing of some novel herbicides for regaining sensitivity. J Mol Model 18:1431–1445
Singh DV, Agarwal S, Kesharwani RK, Misra K (2012) Molecular modelling and computational simulation of the photosystem-II reaction center to address isoproturon resistance in Phalaris minor. J Mol Model 18:3903–3913
Zhang H, Yao Y, Yang H, Wang X, Kang Z, Li Y, Li G, Wang Y (2012) Molecular dynamics and free energy studies on the carboxypeptidases complexed with peptide/small molecular inhibitor: mechanism for drug resistance. Insect Biochem Mol Biol 42:583–595
Liu J, Liu M, Yao Y, Wang J, Li Y, Li G, Wang Y (2012) Identification of novel potential β-N-Acetyl-D-hexosaminidase inhibitors by virtual screening, molecular dynamics simulation and MM-PBSA calculations. Int J Mol Sci 13:4545–4563
Kang T, Gao S, Zhao L, Zhai Y, Ye F, Fu Y (2021) Design, synthesis, and SAR of Novel 1,3-disubstituted imidazolidine or hexahydropyrimidine derivatives as herbicide safeners. J Agric Food Chem 69:45–54
Fu Y, Wang M, Zhao L, Zhang S, Liu Y, Guo Y, Zhang D, Gao S, Ye F (2021) Design, synthesis, herbicidal activity and CoMFA of aryl-formyl piperidinone HPPD inhibitors. Pestic Biochem Physiol 174:104811
Kishore G, Singh DV (2018) Isoproturon tolerance and resistance in Phalaris minor: sequence and structural similarity in Phalaris minor and wheat D1 protein. Pestic Res J 30:246–250
Burgos NR (2015) Whole-plant and seed bioassays for resistance confirmation. Weed Sci 63:152–165
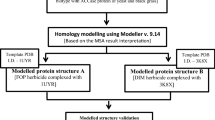
Fiser A, Sali A (2003) Modeller: generation and refinement of homology based protein structure models. Methods Enzymol 374:461–491
Guex N, Peitsch MC (1997) SWISS-MODEL and Swiss-Pdb Viewer: an environment for comparative protein modelling. Electrophoresis 18:2714–2723
Laskowski RA, MacArthur MW, Moss DS, Thornton J (1993) PROCHECK: a program to check the stereo chemical quality of protein structures. J Appl Crystallogr 26:283–291
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35:407–410
Zhang Y, Skolnick J (2005) TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res 33:2302–2309
Irwin JJ, Shoichet BK (2005) ZINC – a free database of commercially available compounds for virtual screening. J Chem Inf Model 45:177–182
Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J, Bryant SH (2015) PubChem substance and compound databases. Nucleic Acids Res 44:202–213
Thomsen R, Christensen MH (2006) MolDock: a new technique for high-accuracy molecular docking. J Med Chem 49:3315–3321
Yuan Y, Pei J, Lai L (2011) LigBuilder 2: a practical de novo drug design approach. J Chem Inf Model 51:1083–1091
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) Autodock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 16:2785–2791
Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447
Schuttelkopf AW, Aalten MFV (2004) PRODRG: a tool for high throughput crystallography of protein-ligand complexes. Acta Crystallogr 60:1355–1363
Marrink SJ, Berger O, Tieleman DP, Jahnig F (1998) Adhesion forces of lipids in a phospholipid membrane studied by molecular dynamics simulations. Biophys J 74:931–943
Tieleman DP, MacCallum JL, Ash WL, Kandt C, Xu Z, Monticelli L (2006) Membrane protein simulations with a united-atom lipid and all-atom protein model: lipid–protein interactions, side chain transfer free energies and model proteins. J Phys: Condens Matter 18:1221
Berendsen HJC, Postma JPM, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690
Oostenbrink C, Villa A, Mark AE, Gunsteren WFV (2004) A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J Comput Chem 25:1656–1676
Amiri S, Sansom MS, Biggin PC (2007) Molecular dynamics studies of AChBP with nicotine and carbamylcholine: the role of water in the binding pocket. Protein Eng Des Sel 20:353–359
Humphrey W, Dalke A, Schulten K (1996) VMD - visual molecular dynamics. J Mol Graph 14:33–38
Kumari R, Kumar R, Open Source Drug Discovery Consortium, Lynn A (2014) g_mmpbsa-A GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54:1951–62
McQuarrie DA (1973) Statistical thermodynamics, 2nd edn. Harper and Row, New York
Broser M, Glockner C, Zouni A (2011) Structural basis of cynobactrial photosystem II inhibition by the herbicides terbutryn. J Biol Chem 286:15964–15972
Sippl MJ (1993) Recognition of errors in three dimensional structures of proteins. Proteins 17:355–362
Lu Q, Wang DS, Chen CS, Hu YD, Chen CS (2005) Structure-based optimization of phenylbutyrate-derived histone deacetylase inhibitors. J Med Chem 48:5530–5553
Acknowledgements
The authors of this manuscript acknowledge Prof. Krishna Mishra, Indian Institute of Information Technology Allahabad, and Prof. R. S. Rathore HOD, Dept. of Bioinformatics, for extending all possible support in the completion of this research work.
Funding
The authors acknowledge the funding supported by the Science and Engineering Research Board, India file No.YSS/2015/001662.
Author information
Authors and Affiliations
Contributions
D. V. Singh conceived and supervised the study. N. K. and P. R. performed the calculations and experiments. N. K. and S. A. interpreted the results and writing of the manuscript is done by all the authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kumar, N., Rani, P., Agarwal, S. et al. 6-Ethoxy-4- N-(2-morpholin-4-ylethyl) -2-N-propan-2-yl-1,3, 5-triazine-2, 4-diamine endows herbicidal activity against Phalaris minor a weed of wheat crop field: An in -silico and experimental approaches of herbicide discovery. J Mol Model 28, 77 (2022). https://doi.org/10.1007/s00894-021-05006-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-021-05006-6