Abstract
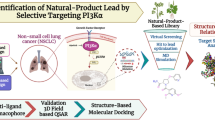
To determine the kinase inhibitory potential of natural products that could be utilized in lung cancer therapy in the near future, a pharmacophore-based activity profiling protocol using parallel pharmacophore-based virtual screening of ZINC—a natural product database—was employed. The work presented here is based on the previously explored fact that pharmacophore-based parallel screening is a reliable in silico protocol to predict the possible biological activities of any compound, or any compound library, by screening it with a number of pharmacophore models. The present study involves ligand-based pharmacophore modeling of various kinases, including EGFR (T790 M), cMET, ErbB2, FGFR and ALK, which are well established targets of normal as well resistant lung cancer. The generated pharmacophore models were then utilized for parallel and cross screening. The profiled molecules for each target were then validated using molecular docking and molecular dynamic simulations. The results show that kinase inhibitory activity profiling of some natural product molecules was successfully achieved.

Pharmacophore and activity profiling of natural products for kinases involved in lung cancer






Similar content being viewed by others
References
Schuster D (2011) 3D pharmacophores as tools for activity profiling. Drug Discov Today Technol 7:e205–e211
Steindl TM, Schuster D, Wolber G, Laggner C, Langer T (2006) High-throughput structure-based pharmacophore modelling as a basis for successful parallel virtual screening. J Comput Aided Mol Des 20:703–715
Meslamani J, Li J, Sutter J, Stevens A, Bertrand H-O, Rognan D (2012) Protein–ligand-based pharmacophores: generation and utility assessment in computational ligand profiling. J Chem Inf Model 52:943–955
Singh PK, Singh H, Silakari O (2016) Kinases inhibitors in lung cancer: from benchside to bedside. Biochim Biophys Acta 1866:128–140
Dagogo-Jack I, Engelman JA, Shaw AT (2016) Overcoming on-target resistance to tyrosine kinase inhibitors in lung cancer. Annu Rev Cancer Biol 1:257–274
Tong M, Seeliger MA (2014) Targeting conformational plasticity of protein kinases. ACS Chem Biol 10:190–200
Singh PK, Silakari O (2017) Chemotherapeutics-resistance “arms” race: an update on mechanisms involved in resistance limiting Egfr inhibitors in lung cancer. Life Sci 186:25–32
Solomon BJ, Mok T, Kim D-W, Wu Y-L, Nakagawa K, Mekhail T, Felip E, Cappuzzo F, Paolini J, Usari T (2014) First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 371:2167–2177
Kinghorn AD, Chin Y-W, Swanson SM (2009) Discovery of natural product anticancer agents from biodiverse organisms. Curr Opin Drug Discov Dev 12:189
Safe S, Kasiappan R (2016) Natural products as mechanism-based anticancer agents: Sp transcription factors as targets. Phytother Res 30:1723–1732
Dassault Systèmes BIOVIA (2015) Discovery studio modeling environment, Release 4. Dassault Systemes, San Diego, CA
Li H, Sutter J, Hoffmann R (2000) HypoGen: an automated system for generating 3D predictive pharmacophore models. Pharmaco Percept Dev Use Drug Design 2:171
Brogi S, Kladi M, Vagias C, Papazafiri P, Roussis V, Tafi A (2009) Pharmacophore modeling for qualitative prediction of antiestrogenic activity. J Chem Inf Model 49:2489–2497
Rubin DB (1980) Randomization analysis of experimental data: the fisher randomization test comment. J Am Stat Assoc 75:591–593
Kirchmair J, Ristic S, Eder K, Markt P, Wolber G, Laggner C, Langer T (2007) Fast and efficient in silico 3D screening: toward maximum computational efficiency of pharmacophore-based and shape-based approaches. J Chem Inf Model 47:2182–2196
Jenkins JL, Kao RY, Shapiro R (2003) Virtual screening to enrich hit lists from high-throughput screening: a case study on small-molecule inhibitors of angiogenin. Prot Struct Funct Bioinform 50:81–93
Hanley JA, McNeil BJ (1982) The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143:29–36
Kalva S, Singam EA, Rajapandian V, Saleena LM, Subramanian V (2014) Discovery of potent inhibitor for matrix metalloproteinase-9 by pharmacophore based modeling and dynamics simulation studies. J Mol Graph Model 49:25–37
Irwin JJ, Shoichet BK (2005) ZINC− a free database of commercially available compounds for virtual screening. J Chem Inf Model 45:177–182
Wu G, Robertson DH, Brooks CL, Vieth M (2003) Detailed analysis of grid-based molecular docking: a case study of CDOCKER—A CHARMm-based MD docking algorithm. J Comput Chem 24:1549–1562
Deshpande N, Addess KJ, Bluhm WF, Merino-Ott JC, Townsend-Merino W, Zhang Q, Knezevich C, Xie L, Chen L, Feng Z (2005) The RCSB protein data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res 33:D233–D237
Singh PK, Silakari O (2018) In-silico guided development of imine based inhibitors for resistance-deriving kinases. J Biomol Struct Dyn:1–21
Singh PK, Silakari O (2017) Novel EGFR (T790M)-cMET dual inhibitors: putative therapeutic agents for non-small-cell lung cancer. Future Med Chem 9:469–483
Maestro (2018) Schrödinger Release 2018-3, LLC, New York, NY
Singh PK, Silakari O (2018) Molecular dynamics guided development of indole based dual inhibitors of EGFR (T790M) and c-MET. Bioorg Chem 79:163–170
Acknowledgment
The authors thank the Department of Biotechnology (DBT), New Delhi for awarding funds and junior research fellowship (J.R.F.); BT/PR8275/BID/7/455/2013.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(DOCX 9441 kb)
Rights and permissions
About this article
Cite this article
Singh, P.K., Silakari, O. Pharmacophore and molecular dynamics based activity profiling of natural products for kinases involved in lung cancer. J Mol Model 24, 318 (2018). https://doi.org/10.1007/s00894-018-3849-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-018-3849-7