Abstract
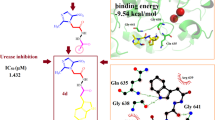
For the latter half of the twentieth century, most medical professionals considered bacterial infection to be a primary cause of gastrointestinal ulcers in human beings. In 1994, the World Health Organization (WHO) recognized Helicobacter pylori, the bacterium most closely linked to ulcer development, as a type I carcinogen. Biological research has shown that there is a positive correlation between the number of species in the Helicobacter genus and the number of medical conditions associated with Helicobacter infection, both of which are increasing rapidly. N-Benzylaniline derivatives, frequently used in industrial manufacturing, are being considered as a strong candidate for ongoing drug modeling in search of novel therapies. The basic goal behind this study was to determine the potency of experimentally proved data, and to determine favorable substituents to enhance potency, and thereafter to support this finding through theoretical modification of the existing base skeleton by addition of suitable substituents. Ligands were investigated thoroughly by paying attention to the urease-inhibitory properties present in the selected series. Initially, docking was performed on ligands with protein to produce efficient docking poses. Molecular dynamics (MD) simulations were also performed to precisely understand the interactions between ligands and proteins. Thereafter, MM-GBSA was used in order to validate the methods and results. Good interaction was observed with amino acids Arg338, Ala169, Asp223, His322, and Asn168. This study also revealed that the electron rich hydroxyl group (–OH) substituent plays an important role during bond formation. In addition, various hydrogen bonds, ionic bonds, and pi–pi stacking bonds make significant contributions towards urease inhibition. Therefore, further research utilizing electron-rich moieties may lead to novel and efficacious urease inhibitors.





























Similar content being viewed by others
References
Marshall BJ (1983) Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet 1:1273–1275
Blaser MJ (1990) Helicobacter pylori and the pathogenesis of gastro duodenal inflammation. J Infect Dis 161:626–633
Kitahara F, Shimazaki R, Sato T, Kojima Y, Morozumi A, Fujino MA (1998) Severe atrophic gastritis with Helicobacter pylori infections and gastric cancer. Gastric Cancer 1:118–124
Kusters JG, Van Vliet AHM, Kuipers EJ (2006) Pathogenesis of Helicobacter pylori infection. Clin Microbial Rev 19:449–490
Shokerzadeh L, Baqhaei K, Yamaoka Y, Dabiri H, Jafari F, Sahebekhtiari N, Tahami A, Suqianoto M, Zoiaji H, Zali MR (2010) Analysis of 3-end variable region of the caga gene in Helicobacter pylori isolated from Iranian population. J Gastroenterol Hepatol 25:172–177
Hołubiuk Ł, Imiela J (2016) Diet and Helicobacter pylori infection. Prz Gastroenterol 11:150–154
Joseph IM, Kirschner D (2004) A model for the study of Helicobacter pylori interaction with human gastric acid secretion. J Theore Bio 228:55–80
H. pylori Transmission and spread of infection. Mel and Enid Zuckerman College of Public Health. Available at: https://publichealth.arizona.edu/outreach/health-literacy-awareness/hpylori/transmission. [Accessed 28 August 2017]
Abdullah MA, Abuo-Rahma GE, Abdelhafez EM (2017) Design, synthesis, molecular docking, anti-Proteus mirabilis and urease inhibition of new fluoroquinolone carboxylic acid derivatives. Bioorg Chem 70:1–11
Kafarski P, Talma M (2018) Recent advances in design of new urease inhibitors: a review. J Adv Res 13:101–112
Hassan STS, Sudomova E (2017) Biological evaluation and molecular docking of protocatechuic acid from Hibiscus sabdariffa L. as a potent urease inhibitor by an ESI-MS based method. Molecule 22:1696
Zaborska W, Krajewska B, Kot M, Karcz W (2007) Quinone-induced inhibition of urease: elucidation of its mechanisms by probing thiol groups of the enzyme. Bioorg Chem 35:233–242
Schaffer JN, Melanie M (2015) Pearson, Proteus mirabilis and urinary tract infection. Microbial Spetr 5. https://doi.org/10.1128/microbiolspec.UTI-0017-2013
Hovelius B, Mardh PA (1984) Staphylococcus saprophyticus as a common cause of urinary tract infections. Rev Infect Dis 6:328–337
Mobley HLT (2001) Urease. In: Mendz GL, Hazell SL (eds) Helicobacter pylori: Physiology and genetics, chapter 16. ASM, Washington DC
Hawtin PR, Delves HT, Newell DG (1991) The demonstration of nickel in the urease of Helicobacter pylori by atomic absorption spectroscopy. FEMS Microbiol Lett 77:51–54
Hausinger RP (1987) Nickel utilization by microorganism. Microbial Rev 51:22–24
Han SY et al. (2003) N-Benzylideneaniline and N-Benzylaniline are potent inhibitors of Lignostilbene-α,β-dioxygenase, a key enzyme in oxidative cleavage of the central double bond of Lignostilbene 18: 279–283
Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M (1983) Charmm, A program for macromolecular energy, minimization and dynamics calculations. J Comput Chem 4:187–217
Cornell WD, Cieplak P, Bayly CL, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA (1995) A second generation force field for the simulation of proteins, nuclic acids, and organic molecules. J Am Chem Soc 117:5179–5197
Ensor KB, Glynn PW (1996) Grid–based simulation and the method of Conditional Least Squares. In: Charnes JM, Morrice DJ, Brunner DT, Swain JJ (eds) Proceedings of the 1996 Winter Simulation Conference. IEEE, Piscataway NJ, pp 325–331
Still WC, Tempczyle A, Hawley RC, Hendrickson T (1990) Semianalytical treatment of salvation for molecular mechanics and dynamics. J Am Chem Soc 16:6127–6129
Schrödinger Release 2017-1 (2017) LigPrep, Schrödinger. LLC, New York, NY
Xiao ZP, She WK et al. (2015) Synthesis and evaluation N-analogs of 1,2-diarylethane as Helicobacter pylori urease inhibition 23: 4508–4513
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242
Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aid Mol Des 27:221–234
Shivakumar D, Williams J, Wu Y, Damm W, Shelley J, Sherman W (2010) Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J Chem Theory Comput 6:1509–1519
Ferrari AM, Egliesposti G, Sqobba M, Rastelli G (2007) Validation of an automated procedure for the prediction of relative free energies of binding on a set of aldose reductase inhibitors. Bioorg Med Chem 24:7865–7877
Kurczab R (2017) The evaluation QM/MM-driven molecular docking combined with MM/GBSA calculations as a halogen bond scoring strategy. Acta Cryst 73:188–194
Alexander DLJ, Tropsha A, Winkler DA (2015) Beware of R2: simple, unambiguous assessment of the prediction accuracy of QSAR and QSPR models. J Chem Inf Model 55:1316–1322
Besalu E, de Julian-Ortiz JV, Poqliani L (2007) Trends and plot methods in MLR studies. J Chem Inf Model 47:751–760
Jain AN (2008) Bias, reporting, and sharing: computational evaluations of docking methods. J Comput Aided Mol Des 22:201–212
Oliviero C, Sandor P (2001) A normalized root-mean-square distance for comparing protein three-dimensional structures. Protein Sci 7:1470–1473
Rui-guang GE, Dong-Xian W, Minq-conq H, Xue-song S (2013) Nickel trafficking system responsible for urease maturation in Helicobacter pylori. World J Gastroenterol 45:8211–8218
Correlation Coefficients (2017) Correlation coefficients. [ONLINE] availability: https://www.andrews.edu/~calkins/math/edrm611/edrm05.htm. [Accessed 31 August 2017]
Xiaoqian Z, Xin L, Xiaodong J, Xiangfei M, Jinghua F (2012) The performance analysis of massively parallel program NAMD on th-1A. In: Zeng D (ed) Advances in information technology and industry applications, vol 136. Springer, Berlin, pp 265–270
National Institute of Diabetes and Digestive and Kidney Diseases (2017) Gastritis. NIDDK. Available at: https://www.niddk.nih.gov/health-information/digestive-diseases/gastritis. [Accessed 28 August 2017]
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure statement
All the authors of this manuscript declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Gupta, S., Bajaj, A.V. Extra precision glide docking, free energy calculation and molecular dynamics studies of 1,2-diarylethane derivatives as potent urease inhibitors. J Mol Model 24, 261 (2018). https://doi.org/10.1007/s00894-018-3787-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-018-3787-4