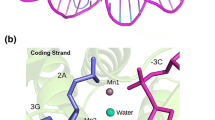
Abstract
BsoBI is a type II restriction endonuclease belonging to the EcoRI family. There is only one previously published X-ray structure for this endonuclease: it shows a homodimer of BsoBI completely encircling DNA in a tunnel. In this work, molecular dynamics simulations were employed to elucidate possible ways in which DNA is loaded into this complex prior to its cleavage. We found that the dimer does not open spontaneously when DNA is removed from the complex on the timescale of our simulations (~ 0.5 μs). A biased simulation had to be used to facilitate the opening, which revealed a possible way for the two catalytic domains to separate. The α-helices connecting the catalytic and helical domains were found to act as a hinge during the separation. In addition, we found that the opening of the BsoBI dimer was influenced by the type of counterions present in the environment. A reference simulation of the BsoBI/DNA complex further showed spontaneous reorganization of the active sites due to the binding of solvent ions, which led to an active-site structure consistent with other experimental structures of type II restriction endonucleases determined in the presence of metal ions.








Similar content being viewed by others
References
Roberts RJ, Macelis D (1998) REBASE—restriction enzymes and methylases. Nucleic Acids Res. 26:338–350. https://doi.org/10.1093/nar/26.1.338
Pingoud A, Fuxreiter M, Pingoud V, Wende W (2005) Type II restriction endonucleases: structure and mechanism. Cell Mol Life Sci CMLS 62:685–707. https://doi.org/10.1007/s00018-004-4513-1
Ruan H, Lunnen K, Pelletier J, Xu S (1997) Overexpression of BsoBI restriction endonuclease in E-coli, purification of the recombinant BsoBI, and identification of catalytic residues of BsoBI by random mutagenesis. Gene 188:35–39. https://doi.org/10.1016/S0378-1119(96)00773-1
van der Woerd M, Pelletier J, Xu S, Friedman A (2001) Restriction enzyme BsoBI-DNA complex: a tunnel for recognition of degenerate DNA sequences and potential histidine catalysis. Structure 9:133–144. https://doi.org/10.1016/S0969-2126(01)00564-0
Zhou XE, Wang Y, Reuter M et al (2004) Crystal structure of type IIE restriction endonuclease EcoRII reveals an autoinhibition mechanism by a novel effector-binding fold. J Mol Biol 335:307–319. https://doi.org/10.1016/j.jmb.2003.10.030
Golovenko D, Manakova E, Tamulaitiene G et al (2009) Structural mechanisms for the 5′-CCWGG sequence recognition by the N- and C-terminal domains of EcoRII. Nucleic Acids Res 37:6613–6624. https://doi.org/10.1093/nar/gkp699
Hashimoto H, Shimizu T, Imasaki T et al (2005) Crystal structures of type II restriction endonuclease EcoO109I and its complex with cognate DNA. J Biol Chem 280:5605–5610. https://doi.org/10.1074/jbc.M411684200
Oroguchi T, Hashimoto H, Shimizu T et al (2009) Intrinsic dynamics of restriction endonuclease EcoO109I studied by molecular dynamics simulations and X-ray scattering data analysis. Biophys J 96:2808–2822. https://doi.org/10.1016/j.bpj.2008.12.3914
Szczepanowski RH, Carpenter MA, Czapinska H et al (2008) Central base pair flipping and discrimination by PspGI. Nucleic Acids Res 36:6109–6117. https://doi.org/10.1093/nar/gkn622
Bochtler M, Szczepanowski RH, Tamulaitis G et al (2006) Nucleotide flips determine the specificity of the Ecl18kI restriction endonuclease. EMBO J 25:2219–2229. https://doi.org/10.1038/sj.emboj.7601096
Viadiu H, Aggarwal AK (1998) The role of metals in catalysis by the restriction endonuclease Bam HI. Nat Struct Mol Biol 5:910–916. https://doi.org/10.1038/2352
Uyar A, Kurkcuoglu O, Nilsson L, Doruker P (2011) The elastic network model reveals a consistent picture on intrinsic functional dynamics of type II restriction endonucleases. Phys Biol 8:056001. https://doi.org/10.1088/1478-3975/8/5/056001
Ehbrecht HJ, Pingoud A, Urbanke C et al (1985) Linear diffusion of restriction endonucleases on DNA. J Biol Chem 260:6160–6166
von Hippel PH, Berg OG (1989) Facilitated target location in biological systems. J Biol Chem 264:675–678
Berman HM, Westbrook J, Feng Z et al (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242. https://doi.org/10.1093/nar/28.1.235
Marti-Renom MA, Stuart AC, Fiser A et al (2000) Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct 29:291–325. https://doi.org/10.1146/annurev.biophys.29.1.291
Eswar N, Webb B, Marti-Renom MA et al (2007) Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci Chap 2:Unit 2.9. https://doi.org/10.1002/0471140864.ps0209s50
Grubmuller H (1996) Solvate 1.0. Ludwig-Maximilians-Universität, München
Case D, Darden T, Cheatham T et al (2012) AMBER 12. University of California, San Francisco
Hornak V, Abel R, Okur A et al (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins Struct Funct Bioinf 65:712–725. https://doi.org/10.1002/prot.21123
Pérez A, Marchán I, Svozil D et al (2007) Refinement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys J 92:3817–3829. https://doi.org/10.1529/biophysj.106.097782
Jorgensen W, Chandrasekhar J, Madura J et al (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79(935):926
Aqvist J (1990) Ion water interaction potentials derived from free-energy perturbation simulations. J Phys Chem 94:8021–8024
Smith DE, Dang LX (1994) Computer simulations of NaCl association in polarizable water. J Chem Phys 100:3757–3766. https://doi.org/10.1063/1.466363
Darden T, York D, Pedersen L (1993) Particle mesh Ewald—an N.log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092. https://doi.org/10.1063/1.464397
Salomon-Ferrer R, Götz AW, Poole D et al (2013) Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J Chem Theory Comput 9:3878–3888. https://doi.org/10.1021/ct400314y
Roe DR, Cheatham TE (2013) PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput 9:3084–3095. https://doi.org/10.1021/ct400341p
Kulhánek P, Štěpán J, Oľha J et al (2016) Conversion and analysis tools. https://lcc.ncbr.muni.cz/whitezone/development/cats/index.html
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14(33–38):27–28
Stone J (1998) An efficient library for parallel ray tracing and animation. Master’s thesis. Computer Science Department, University of Missouri–Rolla, Rolla
Lu X-J, Olson WK (2003) 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res 31:5108–5121. https://doi.org/10.1093/nar/gkg680
Sanner MF, Olson AJ, Spehner JC (1996) Reduced surface: an efficient way to compute molecular surfaces. Biopolymers 38:305–320. https://doi.org/10.1002/(SICI)1097-0282(199603)38:3<305::AID-BIP4>3.0.CO;2-Y
Amadei A, Linssen A, Berendsen H (1993) Essential dynamics of proteins. Proteins Struct Funct Genet 17:412–425. https://doi.org/10.1002/prot.340170408
VanAalten DMF, DeGroot BL, Findlay JBC et al (1997) A comparison of techniques for calculating protein essential dynamics. J Comput Chem 18:169–181
Baker NA, Sept D, Joseph S et al (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci 98:10037–10041. https://doi.org/10.1073/pnas.181342398
Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA (2004) PDB2PQR: an automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res 32:W665–W667. https://doi.org/10.1093/nar/gkh381
Dolinsky TJ, Czodrowski P, Li H et al (2007) PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res 35:W522–W525. https://doi.org/10.1093/nar/gkm276
Konig PH, Ghosh N, Hoffmann M et al (2006) Toward theoretical analyis of long-range proton transfer kinetics in biomolecular pumps. J Phys Chem A 110:548–563. https://doi.org/10.1021/jp052328q
Olsson MHM, Sondergaard CR, Rostkowski M, Jensen JH (2011) PROPKA3: consistent treatment of internal and surface residues in empirical pK(a) predictions. J Chem Theory Comput 7:525–537. https://doi.org/10.1021/ct100578z
Laio A, Parrinello M (2002) Escaping free-energy minima. Proc Natl Acad Sci USA 99:12562–12566. https://doi.org/10.1073/pnas.202427399
Kulhánek P, Štěpán J, Fuxreiter M et al (2013) PMFLib—a toolkit for free energy calculations. https://lcc.ncbr.muni.cz/whitezone/development/pmflib/index.html
Lee J, Chang J, Joseph N et al (2005) MutH complexed with hemi- and unmethylated DNAs: coupling base recognition and DNA cleavage. Mol Cell 20:155–166. https://doi.org/10.1016/j.molcel.2005.08.019
Jin L, Ye F, Zhao D et al (2014) Metadynamics simulation study on the conformational transformation of hhai methyltransferase: an induced-fit base-flipping hypothesis. Biomed Res Int 2014:304563. https://doi.org/10.1155/2014/304563
Formoso E, Limongelli V, Parrinello M (2015) Energetics and structural characterization of the large-scale functional motion of adenylate kinase. Sci Rep 5:srep08425. https://doi.org/10.1038/srep08425
Pace CN, Scholtz JM (1998) A helix propensity scale based on experimental studies of peptides and proteins. Biophys J 75:422–427
Acknowledgements
This research was carried out under the project CEITEC 2020 (LQ1601) with financial support from the Ministry of Education, Youth and Sports of the Czech Republic under the National Sustainability Programme II. Computational resources were provided by CESNET LM2015042, CERIT Scientific Cloud LM2015085, and the IT4Innovations National Supercomputing Center LM2015070, as provided under the program “Large Infrastructures for Research, Experimental Development and Innovations” by the Ministry of Education, Youth and Sports.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Dedicated to Dr. Peter Politzer on the occasion of his 80th birthday.
This paper belongs to Topical Collection P. Politzer 80th Birthday Festschrift
Electronic supplementary material
ESM 1
(PDF 3973 kb)
Rights and permissions
About this article
Cite this article
Štěpán, J., Kabelka, I., Koča, J. et al. Behavior of BsoBI endonuclease in the presence and absence of DNA. J Mol Model 24, 22 (2018). https://doi.org/10.1007/s00894-017-3557-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-017-3557-8