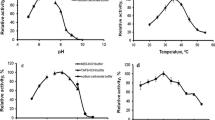
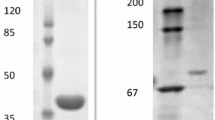
Abstract
Four enzymes involved in sucrose metabolism: sucrose phosphate synthase (Sps), sucrose phosphate phosphatase (Spp), sucrose synthase (Sus) and fructokinase (FruK), were obtained as his-tagged proteins from the moderately thermophilic methanotroph Methylocaldum szegediense O12. Sps, Spp, FruK and Sus demonstrated biochemical properties similar to those of other bacterial counterparts, but the translated amino acid sequences of Sps and Spp displayed high divergence from the respective microbial enzymes. The Sus of M. szegediense O12 catalyzed the reversible reaction of sucrose cleavage in the presence of ADP or UDP and preferred ADP as a substrate, thus implying a connection between sucrose and glycogen metabolism. Sus-like genes were found only in a few methanotrophs, whereas amylosucrase was generally used in sucrose cleavage in this group of bacteria. Like other microbial fructokinases, FruK of M. szegediense O12 showed a high specificity to fructose.



Similar content being viewed by others
References
Akutsu J, Zhang Z, Morita R, Kawarabayasi Y (2015) Identification and characterization of a thermostable bifunctional enzyme with phosphomannose isomerase and sugar-1-phosphate nucleotidylyltransferase activities from a hyperthermophilic archaeon, Pyrococcus horikoshii OT3. Extremophiles 19(6):1077–1085
Bodrossy L, Holmes EM, Holmes AJ, Kovacs KL, Murrell JC (1997) Analysis of 16S rRNA and methane monooxygenase gene sequences reveals a novel group of thermotolerant and thermophilic methanotrophs, Methylocaldumgen. nov. Arch Microbiol 168:493–503
Bruneau JM, Worrell AC, Cambou B, Lando D, Voelker TA (1991) Sucrose phosphate synthase, a key enzyme for sucrose biosynthesis in plants. Plant Physiol 96:473–478
But SY, Rozova ON, Khmelenina VN, Reshetnikov AS, Trotsenko YA (2012) Properties of recombinant ATP dependent fructokinase from the halotolerant methanotroph Methylomicrobium alcaliphilum 20Z. Biochemistry (Moscow) 77:372–377
But SY, Khmelenina VN, Reshetnikov AS, Trotsenko YA (2013a) Bifunctional sucrose phosphate synthase/phosphatase is involved in the sucrose biosynthesis by Methylobacillus flagellatus KT. FEMS Microbiol Lett 347:43–51
But SY, Khmelenina VN, Reshetnikov AS, Trotsenko YA (2013b) Construction and characterization of Methylomicrobium alcaliphilum 20Z knockout mutants defective in sucrose and ectoine biosynthesis genes. Microbiology (Moscow) 82(2):253–255
But SY, Khmelenina VN, Reshetnikov AS, Mustakhimov II, Kalyuzhnaya MG, Trotsenko YA (2015) Sucrose metabolism in halotolerant methanotroph Methylomicrobium alcaliphilum 20Z. Arch Microbiol 197(3):471–480
Caescu C, Vidal O, Krzewinski O, Artenie V, Bouquelet S (2004) Bifidobacterium longum requires a fructokinase (Frk; ATP:d-fructose-6-phosphotransferase, EC 2.7.1.4) for fructose catabolism. J Bacteriol 186:6515–6525
Cumino A, Ekeroth C, Salerno GL (2001) Sucrose-phosphate phosphatase from Anabaena sp. strain PCC 7120: isolation of the protein and gene revealed significant structural differences from the higher-plant enzyme. Planta 214(2):250–256
Cumino AC, Marcozzi C, Barreiro R, Salerno GL (2007) Carbon cycling in Anabaena sp. PCC 7120. Sucrose synthesis in the heterocysts and possible role in nitrogen fixation. Plant Phys 143:1385–1397
Curatti L, Desplats EFP, Abratti G, Limones V, Herrera-Estrella L, Salerno G (1998) Sucrose-phosphate synthase from Synechocystis sp. PCC 6803: identification of the spsA gene and characterization of the enzyme expressed in Escherichia coli. J Bacteriol 180:6776–6779
Curatti L, Giarrocco LE, Cumino AC, Salerno GL (2008) Sucrose synthase is involved in the conversion of sucrose to polysaccharides in filamentous nitrogen-fixing cyanobacteria. Planta 228:617–625
Diricks M, De Bruyn F, Van Daele P, Walmagh M, Desmet T (2015) Identification of sucrose synthase in nonphotosynthetic bacteria and characterization of the recombinant enzymes. Appl Microbiol Biotechnol 99:8465–8474
Ehira S, Kimura S, Miyazaki S, Ohmori M (2014) Sucrose synthesis in the nitrogen-fixing Cyanobacterium Anabaena sp. strain PCC 7120 is controlled by the two-component response regulator OrrA. Appl Environ Microbiol 80:5672–5679
Eshinimaev BTS, Medvedkova KA, Khmelenina VN, Suzina NE, Osipov GA, Lysenko AM, TrotsenkoIu A (2004) New thermophilic methanotrophs of the genus Methylocaldum. Mikrobiologiia (Moscow) 73:530–539
Figueroa CM, AsenciónDiez MD, Kuhn ML, McEwen S, Salerno GL, Iglesias AA, Ballicora MA (2013) The unique nucleotide specificity of the sucrose synthase from Thermosynechococcus elongatus. FEBS Lett 587:165–169
Islam T, Larsen Ø, Torsvik V, Øvreås L, Panosyan H, Murrell JC, Birkeland N-K, Bodrossy L (2015) Novel methanotrophs of the family Methylococcaceae from different geographical regions and habitats. Microorganisms 3:484–499
Khmelenina VN, Kalyuzhnaya MG, Sakharovsky VG, Suzina NE, Trotsenko YA, Gottschalk G (1999) Osmoadaptation in halophilic and alkaliphilicmethanotrophs. Arch Microbiol 172:321–329
King K, Phan P, Rellos P, Scopes RK (1996) Overexpression, purification, and generation of a thermostable variant of Zymomonas mobilis fructokinase. Protein Expr Purif 7:373–376
Klähn S, Hagemann M (2011) Compatible solute biosynthesis in cyanobacteria. Environ Microbiol 13:551–562
Klotz KL, Finger FL, Shelver WL (2003) Characterization of two sucrose synthase isoforms in sugar beet root. Plant Physiol Biochem 41:107–115
Kolman MA, Torres LL, Martin ML, Salerno GL (2012) Sucrose synthase in unicellular cyanobacteria and its relationship with salt and hypoxic stress. Planta 235:955–964
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(− Delta Delta C(T)) method. Methods 25:402–408
Lunn J (2002) Evolution of sucrose synthesis Plant Physiol 128:1490–1500
Lunn JE, Price GD, Furbank RT (1999) Cloning and expression of a prokaryotic sucrose-phosphate synthase gene from the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol 40:297–305
Martínez-Noël GM, Cumino AC, KolmanMde L, Salerno GL (2013) First evidence of sucrose biosynthesis by single cyanobacterial bimodular proteins. FEBS Lett 587:1669–1674
Medvedkova KA, Khmelenina VN, Trotsenko YA (2007) Sucrose as a factor of thermal adaptation of the thermophilic methanotroph Methylocaldum szegediense O-12. Mikrobiologiya (Moscow) 76:500–502
Medvedkova KA, Khmelenina VN, Baskunov BP, TrotsenkoIu A (2008) Synthesis of melanine by a moderately thermophilic methanotroph Methylocaldum szegediense O-12 depends on cultivation temperature. Mikrobiologiya (Moscow) 77:126–128
Murao S, Nakatani A, Kaneda N (1995) Isolation and characterization of fructokinase from Pseudomonas sp. KN-21. Biosci Biotechnol Biochem 59:1798–1800
Porchia AC, Curatti L, Salerno GL (1999) Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably different from the plant enzymes with respect to substrate affinity and amino-terminal sequence. Planta 210:34–40
Reshetnikov AS, Khmelenina VN, Trotsenko YA (2006) Characterization of the ectoine biosynthesis genes of haloalkalotolerant obligate methanotroph “Methylomicrobium alcaliphilum 20Z”. Arch Microbiol 184:286–297
Rozova ON, Khmelenina VN, Vuilleumier S, Trotsenko YA (2010) Characterization of recombinant pyrophosphate-dependent 6-phosphofructokinase from halotolerant methanotroph Methylomicrobium alcaliphilum20Z. Res Microbiol 161:861–868
Salerno GL, Curatti L (2003) Origin of sucrose metabolism in higher plants: when, how and why? Trends Plant Sci 8:63–69
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, New York
Sinha AK, Pathre U, Sane PV (1997) Purification and characterization of sucrose-phosphate synthase from Prosopis juliflora. Phytochemistry 46:441–447
Slater GG (1969) Stable pattern formation and determination of molecular size by pore-limit electrophoresis. Anal Chem 41(8):1039–1041
Thomson JD, Gibson TJ, Plewniak Jeanmougin F, Higgins DG (1997) The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res 24:4876–4882
Torres LL, Salerno GL (2007) A metabolic pathway leading to mannosylfructose biosynthesis in Agrobacterium tumefaciens uncovers a family of mannosyltransferases. Proc Natl Acad Sci USA 104:14318–14323
Winter H, Huber S (2000) Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes. Crit Rev Plant Sci 19:31–67
Wu B, Zhang Y, Zheng R, Guo C, Wang PG (2002) Bifunctional phosphomannose isomerase/GDP-d-mannose pyrophosphorylase is the point of control for GDP-d-mannose biosynthesis in Helicobacter pylori. FEBS Lett 519:87–92
Yang Y, Shan J, Zhang J, Zhang X, Xie S, Liu Y (2014) Ammonia- and methane-oxidizing microorganisms in high-altitude wetland sediments and adjacent agricultural soils. Appl Microbiol Biotechnol 98:10197–10209
Yen SF, Su JC, Sung HY (1994) Purification and characterization of rice sucrose synthase isozymes. Biochem Mol Biol Int 34:613–620
Acknowledgements
The work was supported by the Russian Foundation for Basic Research #16-04-00462-a and by the Russian Scientific Foundation #18-14-00326. The authors are grateful to all members of the Organization for Methanotroph Genome Analysis for their collaboration (OMeGA), the U.S. Department of Energy Joint Genome Institute and Genoscope for the access to methanotrophic genomes for comparative analyses.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by F. Robb.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
But, S.Y., Solntseva, N.P., Egorova, S.V. et al. The genes and enzymes of sucrose metabolism in moderately thermophilic methanotroph Methylocaldum szegediense O12. Extremophiles 22, 433–445 (2018). https://doi.org/10.1007/s00792-018-1006-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-018-1006-y