Abstract
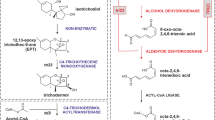
Trichothecenes are the secondary metabolites produced by Trichoderma spp. Some of these molecules have been reported for their ability to stimulate plant growth by suppressing plant diseases and hence enabling Trichoderma spp. to be efficiently used as biocontrol agents in modern agriculture. Many of the proteins involved in the trichothecenes biosynthetic pathway in Trichoderma spp. are encoded by the genes present in the tri cluster. Tri4 protein catalyzes three consecutive oxygenation reaction steps during biosynthesis of isotrichodiol in the trichothecenes biosynthetic pathway, while tri11 protein catalyzes the C4 hydroxylation of 12, 13-epoxytrichothec-9-ene to produce trichodermol. In the present study, we have homology modelled the three-dimensional structures of tri4 and tri11 proteins. Furthermore, molecular dynamics simulations were carried out to elucidate the mechanism of their action. Both tri4 and tri11 encode for cytochrome P450 monooxygenase like proteins. These data also revealed effector-induced allosteric changes on substrate binding at an alternative binding site and showed potential homotropic negative cooperativity. These analyses also showed that their catalytic mechanism relies on protein–ligand and protein–heme interactions controlled by hydrophobic and hydrogen-bonding interactions which orient the complex in optimal conformation within the active sites.






Similar content being viewed by others
Abbreviations
- MD:
-
Molecular dynamics
- TDN:
-
Trichodiene
- EPT:
-
12, 13-epoxytrichothec-9-ene
- SMs:
-
Secondary metabolites
- SRSs:
-
Substrate recognition sites
- P450s:
-
Cytochrome P450 monooxygenases
References
Reino JL, Guerrer ORF, Hrnndez-Galn RIG, Collado IG (2008) Phytochem Rev 7:89–123
Hermosa R, Viterbo A, Chet I, Monte E (2012) Microbiology 158:17–25
Vinale F, Sivasithamparam K, Ghisalberti EL, Marra R, Barbetti MJ, Li H, Woo SL, Lorito M (2008) Physiol Mol Plant Pathol 72:80–86
Vinale F, Sivasithamparam K, Ghisalberti EL, Marra R, Woo SL, Lorito M (2008) Soil Biol Biochem 40:1–10
Chen W, Lee MKC, Jefcoate CS-C, Kim SC, Chen FYuJH (2014) Genome Biol Evol 6:1620–1634
Bowen GD, Rovira AD (1999) Adv Agron 66:1–102
Jain A, Singh A, Singh S, Singh HB (2013) J Plant Growth Regul 32:388–398
Malmierca MG, Cardoza RE, Alexander NJ, Mccormick SP, Hermosa RE, Monte E, Gutiérrez S (2012) Appl. Environ Microbiol 78:4856–4868
Cundliffe E, Cannon M, Davies J (1974) Proc Natl Acad Sci USA 71:30–34
Cardoza RE, Malmierca MG, Hermosa MR, Alexander NJ, Mccormick SP, Proctor RH, Tijerino AM, Rumbero A, Monte E, Gutie S (2011) Appl. Environ Microbiol 77:4867–4877
Degenkolb T, Dieckmann R, Nielsen KF, Gräfenhan T, Theis C, Zafari D, Chaverri P, Ismaiel A, Brückner H, Döhren HV (2008) Mycol Prog 7:177–219
Tijerino A, Cardoza RE, Moraga J, Malmierca Vicente F, Aleu J, Collado IG, Gutiérrez S, Monte S, Hermosa R (2011) Fungal Genet. Biol 48:285–296
Shentu XP, Yuan XF, Liu WP, Xu JF, Yu XP (2015) Am J Biochem Biotechnol 11:169
Hermosa R, Rubio MB, Cardoza RE, Nicolas C, Monte E, Gutiérrez S (2013) Int Microbiol 16:69–80
Malmierca MG, Izquierdo I, Bueno McCormick SP, Cardoza RE, Alexander NJ, Barua J, Lindo L, Casquero PA, Collado IG, Monte E (2016) Environ. Microbiol 18:3991–4004
Pusztahelyi T, Holb IJ, Pocsi I (2015) Front Plant Sci 6:573
Kumari I, Ahmed M, Akhter Y (2016) Int J Biochem Cell Biol 78:370–376
Hlavica P (2017) J Inorg Biochem 167:100–115
Cresnar B, Petric S (2011) Biochim Biophys Acta (BBA)-Proteins Proteom 1814:29–35
Gotoh O (1992) J Biol Chem 267:83–90
Seifert A, Pleiss J (2009) Proteins Struct Funct Bioinform 74:1028–1035
McGuffin LJ, Bryson K, Jones DT (2000) Bioinformatics 16:404–405
Corpet F (1988) Nucleic Acids Res 16:10881–10890
Kim S, Thiessen PA, Bolton EE, Bryant SH (2015) Nucleic Acids Res 43(W1):W605–W611
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) Nat Protoc 10:845–858
Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A (2006) Curr Protoc Bioinformatics. doi:10.1002/0471250953.bi0506s15
Krieger E, Nabuurs SB, Vriend G (2003) Methods Biochem Anal 44:509–524
Berendsen HJC, Spoel Van Der D, Drunen Van R (1995) Comput Phys Commun 91:43–56
Sandhu P, Akhter Y (2016) Arch Biochem Biophys 592:38–49
Guengerich FP (2001) Chem Res Toxicol 14:611–650
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) J Comput Chem 30:2785–2791
SchuÈttelkopf AW, Aalten Van DMF (2004) Acta Crystallogr Sect D Biol Crystallogr 60:1355–1363
Vohra S, Musgaard M, Bell SG, Wong L, Zhou W, Biggin PC (2013) Protein Sci 22:1218–1229
Oostenbrink C, Villa A, Mark AE, Van Gunsteren WF (2004) J Comput Chem 25:1656–1676
Micaelo NM, Macedo AL, Goodfellow BJ, Felix V (2010) J Mol Graph Model 29:396–405
Cojocaru XYUV, Mustafa G, Salo Ahen OMH, Lepesheva GI, Wade RC (2015) J Mol Recognit 28:59–73
Graaf CDE, Oostenbrink C, Keizers PHJ, Tvander WIJST, Jongejan A, Vermeulen NPE (2006) J Med Chem 49:2417–2430
Podust LM, Ouellet H, Kries von JP, Montellano de PRO (2009) J Biol Chem 284:25211–25219
Zhang H, Gay SC, Shah M, Foroozesh M, Osawa J, Liu Y, Zhang Q, Stout CD, Halpert JR, Hollenberg PF (2013) Biochemistry 52:355
Guengerich FP, Munro AW (2013) J Biol Chem 288:17065–17073
Bischoff R, Schlüter H (2012) J Proteom 75:2275–2296
Hasemann CA, Kurumbail RG, Boddupalli SS, Peterson JA, Deisenhofer J (1995) Structure 3:41–62
Guallar V, Baik MH, Lippard SJ, Friesner RA (2003) Proc Natl Acad Sci 100:6998–7002
Podust LM, Sherman DH (2012) Nat Prod Rep 29:1251–1266
Zanger UM, Schwab M (2013) Pharmacol Ther 138:103–141
Kimura M, Tokai T, Takahashi N, Ohsato S (2007) Biosci Biotechnol Biochem 71:2105–2123
Acknowledgements
We acknowledge Central University of Himachal Pradesh and Bioinformatics Resources and Applications Facility, Centre for Development in Advanced Computing, Pune for providing the computational infrastructure. RH acknowledges National Fellowship for Higher Education from University Grants Commission, Govt. of India (UGC). SS receives research stipend from UGC. Research in YA lab is supported by extramural research funds from UGC, Science and Engineering Research Board (DST, Govt. of India), and Indian Council of Medical Research. We thank Dr. P. Aparoy for his generous help during the revision. Prof. Claudio Luchinat (editor-in-chief) and two anonymous referees are also sincerely acknowledged, whose insightful comments and advice during the editorial review helped us to improve our work enormously.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hussain, R., Kumari, I., Sharma, S. et al. Catalytic diversity and homotropic allostery of two Cytochrome P450 monooxygenase like proteins from Trichoderma brevicompactum . J Biol Inorg Chem 22, 1197–1209 (2017). https://doi.org/10.1007/s00775-017-1496-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-017-1496-6