Abstract
Bacteria overexpress, under condition of starvation or oxidative stress, Dps (DNA-binding proteins from starved cells), hollow sphere formed by 12 identical subunits endowed with ferritin-like activity. The iron oxidation and incorporation in Dps take place using H2O2 produced under starvation as preferred iron oxidant, thereby protecting bacteria from oxidative damage. Even if the role of Dps is well known, the mechanism of iron oxidation and incorporation remain to be elucidated. Here, we have used the EPR technique to shed light on the Fe(II) binding and oxidation mechanism at the ferroxidase center using both the wild-type (wt) protein and mutants of the iron ligands (H31G, H43G and H31G-H43G-D58A). The EPR titration of wt Dps and the H31G mutant with Fe(II) upon H2O2 addition shows that Fe(II) is oxidized with the increase of the signal at g = 4.3, reaching a maximum for 12 Fe(II)/subunit. The EPR signal becomes negligible when the titration is carried out on the triple mutant. These experiments indicate that the iron firstly occupied the A site at the ferroxidase center and confirm that the residues H31, H43 and D58 have a key role in the iron oxidation and incorporation process. Moreover, the data indicate that the ferroxidase center, upon mutation of H31 or H43 to Gly, changes the mode of iron binding. Finally, we demonstrate here that, when the iron micelle forms, the EPR signal at g = 4.3 disappears indicating that iron leaves the ferroxidase center to reach the inner cavity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Iron is essential for most living organisms since it serves as a cofactor in several enzymes and as a catalyst in electron transfer processes. However, iron is poorly available and potentially toxic, since Fe(III), the stable oxidation state of the metal at neutral pH values, forms insoluble hydroxy-aquo complexes, while Fe(II) reacts with oxygen forming reactive oxygen species (ROS) through the Fenton reaction:
Hydroxyl radicals and hydrogen peroxide products in this reaction can damage both DNA, proteins and membrane lipids. All organisms therefore have developed strategies that allow them to acquire iron, and to solubilize and store it in a non-toxic, readily available form.
The eukaryotes and bacteria use the same general strategies to counteract the reactivity of iron, i.e., iron is sequestered by ferritins and ROS are neutrilized by specific enzymes as superoxide dismutases and catalase. In 1992, a new protein family which possesses iron storage and detoxification ability was identified in bacteria and named Dps (DNA-binding proteins from starved cells) from the prototype expressed in Escherichia coli under starvation or oxidative stress [1]. Dps proteins were found to protect DNA from oxidative damage, an ability initially attributed to their capacity to bind to DNA without apparent sequence specificity. Other studies have shown successively that Dps proteins possess a ferritin-like activity allowing them to oxidize and incorporate iron and to reduce hydrogen peroxide. This discovery stems to a large extent from the work on Listeria Dps that was initially identified as mini-ferritin, since it was able to oxidize and incorporate iron like an actual ferritin [2].
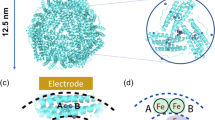
The resolution of the X-ray crystal structure of Listeria Dps at 2.35 Å resolution disclosed that the protein shares all the features of the Dps protein family [3]. Indeed, it is a very stable dodecamer formed by 12 identical subunits assembled with a 23 symmetry (Fig. 1a) just like E. coli Dps, the family prototype [4, 5]. More importantly, the X-ray structure revealed the presence of a very unusual ferroxidase site that is not located in the four-helix bundle of individual monomers as in the 24-mer ferritin, but it is placed at the interface between twofold symmetry-related monomers with iron ligands provided by both the subunits (Fig. 1b). The structural analysis of Listeria Dps shows that iron is coordinated by Glu 62 and Asp 58, located on the B helix (Fig. 1c) of one subunit; by His 31, located on the A helix of the twofold symmetry-related monomer; and by a water molecule at ~ 3 Å from the iron, forming a hydrogen bond with a second histidine, His 43, from the same subunit. These residues are conserved in the Dps proteins, suggesting that iron binding could be part of the protective effect exerted by Dps proteins on DNA [3]. Over the last years, a number of studies have confirmed that the ferritin-like activity is a characteristic of all Dps proteins, whereas the ability to bind DNA is not shared by all the members of the family due to the variability of the N-terminus, the E. coli protein participates in the interaction by means of three lysine residues (lysines 5, 8, 10, E. coli Dps numbering), placed on the flexible N-terminal tail, which are lacking in other Dps [4]. Indeed, when these residues are mutated, the ability to bind DNA of E. coli Dps dramatically decreases [5].
X-ray structure of Listeria innocua Dps protein: a dodecameric assembly (view along the twofold symmetry axis), b functional dimer, c blowup of the ferroxidase center. The A and B monomers are colored salmon and blue, respectively. The iron ions are represented as sphere and colored orange, and the water molecules bound to His43 are represented as nb-sphere and colored red. The residues of the ferroxidase center are represented as sticks. The residues of the ferroxidase center are indicated and represented as sticks (color figure online)
The chemistry of iron oxidation and deposition has been characterized in E. coli Dps [6] and it appears to be different with respect to the other members of the ferritin superfamily. Indeed, while ferritins in general use O2 as iron oxidant with the production of hydrogen peroxide [7] and bacterioferritins can employ both O2 and H2O2 [8], Dps proteins typically use H2O2 as iron oxidant which is usually about 100-fold more efficient than O2 in carrying out iron oxidation [9,10,11]. Zhao and coworkers discover that two Fe(II) may bind at each of the 12 ferroxidase sites and the oxidation of Fe(II) is carried out most efficiently by H2O2 rather than by O2, thereby avoiding hydroxyl radical production through Fenton chemistry [6]. In summary, Dps can acquire up to 500 Fe(III)/dodecamer conserved as oxyhydroxide ferric mineral core according to the mineralization reaction:
In line with the consumption of hydrogen peroxide, spin trapping experiments have indicated that Dps inhibits hydroxyl radical production thereby avoiding DNA damage [3, 6]. The data obtained thereafter on a number of bacterial species brought out clearly the unique capacity of Dps proteins to combat the Fe(II) and H2O2-dependent oxidative stress even though different situations may be encountered in the various species due to the expression of different defense proteins.
The role of the ferroxidase center in iron uptake and hydrogen peroxide detoxification was further investigated by Ilari et al. [12] in Listeria innocua Dps. In this study, the authors substituted the iron ligands His31, His43, and Asp58 with glycine or alanine residues either individually or in combination. The X-ray crystal structures of the variants display small differences in the positions of the ferroxidase site residues compared to the native protein. Fluorimetric titrations underlined that substitution of either His31 or His43 decreases Fe(II) affinity significantly with respect to wt Listeria innocua Dps (K = 105 vs 107 M−1) without altering binding stoichiometry (12 Fe(II)/dodecamer), whereas the H31G-H43G and H31G-H43G-D58A variants do not bind Fe(II). Oxidation of protein-bound Fe(II) with hydrogen peroxide seems to increase the ferroxidase center binding stoichiometry to 24 Fe(III)/dodecamer in the wt protein and the H31G mutant, whereas in H43G, H31GH43G and H31G-H43G-D58A mutants, the fluorescence quenching upon addition of Fe(II) and H2O2 decreases dramatically and is negligible in the triple mutant, indicating that iron does not bind anymore to the ferroxidase center (close to the chromophores W32 and W144). These findings indicate that the ferroxidase center mutations impair the Fe(II) binding and oxidation process and strongly decrease the ability of Dps to protect DNA from oxidative damage in the presence of Fe(II) and hydrogen peroxide [12]. Moreover, while with oxygen as iron oxidant, the ferroxidase site mutations have little or no effect on the kinetics of iron uptake and in the formation of micelles inside the protein shell, they strongly affect, in the presence of hydrogen peroxide as iron oxidant, the rate of iron oxidation and the capacity of Dps to inhibit Fenton chemistry, thereby protecting DNA from oxidative damage.
In the present study, we investigated by EPR spectroscopy the stoichiometry and the mechanism of Fe(II) binding, oxidation, in Listeria Dps and in the site-directed variants of the ferroxidase center.
EPR data confirm that the residues H31, H43 and D58 have a key role in the iron oxidation and incorporation process and allow us to propose a catalytic mechanism for detoxification by Dps proteins.
2 Materials and Methods
2.1 Plasmid Construction
Recombinant wt Listeria innocua Dps was overexpressed in E. coli strain BL21(DE3). The Listeria innocua Dps gene, termed ferritin from Listeria innocua (fri) [13], was obtained by means of PCR techniques using pTZ35 as template and primers A1 (forward primer) and B2 (reverse primer) containing the cleavage sites for restriction enzymes NdeI and BamHI [12]. The resulting 468-bp product was doubly digested with NdeI and BamHI, isolated using the QIAquick gel extraction kit (Qiagen), and subcloned into the expression vector pET-11a (Novagen).
The H31G, H43G, H31G-H43G and H31G-H43G-D58A mutants were generated by the PCR technique with the wt fri gene as template as described by Ilari et al. [12]. PCR reactions were performed by the Qiagen method using Pfu Turbo DNA polymerase (Stratagene). All PCR fragments were digested with NdeI and BamHI and cloned into pET-11a. All products were purified with the Qiagen kit; the purified DNA was transformed into E. coli strain BL21(DE3). The variant constructs were confirmed by DNA sequencing.
2.2 Protein Purification and Characterization
The recombinant (wt) protein and the site-specific mutants were purified as previously described [2, 12]. Briefly, bacterial cells were suspended in 50 mM Tris–HCl, pH 7.4, containing 1 mM EDTA, 0.5 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride and disrupted in a French press. The plasma membranes were removed by centrifugation at 15,000 rpm for 40 min, Then, the supernatant was heated at 70 °C for 10 min and rapidly cooled and centrifuged (15,000 rpm for 30 min) to eliminate unfolded proteins. The supernatant was treated with ammonium sulfate (80% w/v) and the precipitate was collected by centrifugation (10,000 rpm for 20 min), dissolved in 5 ml of 20 mM Tris–HCl, pH 7.4, and dialyzed against the same buffer. To remove DNA, the material was treated with a solution of streptomycin sulfate (1% w/v) for 30 min at room temperature and centrifuged (15,000 rpm for 15 min). The protein was further purified using two chromatographic steps: (i) fast protein liquid chromatography (FPLC) using a Mono-Q column (Pharmacia Biotech Inc.) equilibrated with 20 mM Tris–HCl, pH 7.4, (the protein was eluted with a linear gradient of 0.1–0.3 M NaCl in the same buffer); (ii) ion exchange chromatography on a DEAE-52 column equilibrated with 20 mM Tris–HCl buffer at pH 7.8 (the protein was eluted with a 0.1–0.3 M NaCl gradient).
The wt protein contains 1.8–2.0 Fe/dodecamer as assessed by atomic absorption spectroscopy or the ferrozine method (Fluka), whereas the variants are iron free. Protein concentration was determined spectrophotometrically on the basis of the molar (Mr 216 kDa) absorption coefficient, ε = 2.59 × 105 M−1 cm−1 at 280 nm [2].
2.3 Aerobic Listeria innocua Dps Sample Preparation
The protein solution was transferred into a 1.5 ml Eppendorf tube. The freshly prepared Fe(NH4)2(SO4)2 anaerobic solution was added by a micro-syringe and gently mixed. Samples were incubated in air for 2 days. The solution was transferred by a Gilson pipette to the EPR tube, and frozen with liquid N2 before insertion in the EPR spectrometer at 10 K.
2.4 Anaerobic Listeria innocua Dps Sample Preparation
The protein solution was transferred into a small tube sealed with a rubber cap. Oxygen was removed by bubbling N2 for 30 min with a capillary. A freshly prepared Fe(NH4)2(SO4)2 anaerobic solution, which was prepared flushing for at least 30 min with N2, was added by a micro-syringe and gently mixed. After 2 min, H2O2 was added to the Dps–Fe(II) complex in a Fe(II)/H2O2 ratio of 2/1, either stepwise or in a unique step. The addition of H2O2 was subsequent to avoid its disproportionation. After 2 min, the solution was transferred by a Gilson pipette to the EPR tube and frozen instantaneously with liquid N2 before insertion in the EPR spectrometer at 10 K.
2.5 EPR Measurements
The EPR sample concentration was ~ 0.1 mM for wt Listeria Dps and the site-directed mutants.
Continuous-wave (CW) X-band EPR measurements were performed on a Bruker Elexsys E580 spectrometer at 9.4 GHz, equipped with standard TE102 cavity. The temperature was controlled with a helium flow cryostat (Oxford Instruments ESR-900) driven by a temperature controller. Experimental conditions were: T = 10 K, microwave power = 600 µwatts, field modulation = 100 kHz, modulation amplitude = 10 G, time constant = 82 ms, conversion time = 328 ms.
g values were estimated by calibration using a sample of strong pitch [14].
Spin quantification of the g = 4.3 signal was performed by double integration [15] using as quantification standard the apoprotein, whose iron concentration (1.8 iron/dodecamer for wt Listeria Dps) was independently determined by atomic absorption and ferrozine assay [12].
3 Results
Low-temperature CW-EPR measurements of paramagnetic species were undertaken to assess whether mononuclear haem and non-haem Fe(III) species [16, 17], mixed-valence Fe(II)–Fe(III) complexes [18, 19], or radical species [20, 21] are formed during iron oxidation in the native DPS from Listeria innocua and in different mutants where histidine residues are replaced by glycine. For all these variants, the apoprotein and the protein at different stages of iron uptake do not show any signal in the g = 2 region: the presence of stable mixed-valence Fe(II)–Fe(III) complexes can be excluded, since the characteristic EPR signal at g′ = 1.87 was not observed and narrow signals due to long-lived radical species have also not been detected in any of the samples. For this reason, the focus in the next paragraphs is on the g = 4.3 region, where the signal with isotropic character due to the transition occurring within one of the Kramer doublets of the high-spin mononuclear Fe(III) species (S = 5/2) can be found [22, 23].
Aerobic and anaerobic titrations of native Listeria innocua ferritin were carried out on apoprotein samples adding increasing amount of Fe(II). The anaerobic titration was carried adding together with Fe(II) a stoichiometric amount of H2O2. Figure 2 shows the EPR signal in the region around g = 4.3 as observed in the apoprotein and in the loaded samples. The g = 4.3 signal can be ascribed to a high-spin mononuclear Fe(III) species in an environment of low symmetry. To assess the amount of EPR-detectable iron in the protein samples, the g = 4.3 signal was compared with that of the apoprotein which is used as an intensity standard. Quantification by atomic absorption spectroscopy and the ferrozine assay of the apoprotein gives a total iron content of ~ 1.5 Fe(III)/dodecamer (see Materials and methods). This iron is in the Fe(III) state as proved by the absence of any effect on the EPR signal of the “apoprotein” by addition of H2O2 (see Fig. S1). The most intense signal accounts for almost complete occupancy, ~ 11.5 Fe(III)/dodecamer, after the addition of 12 Fe(II) in the presence of H2O2. For the aerobic titration, the EPR signal was recorded after 2 days from the addition of Fe(II) to the protein sample, but the maximum occupancy is lower and corresponds to 7.0 Fe(III)/dodecamer. Since hydrogen peroxide is a more efficient oxidant for iron in Dps proteins under physiological conditions, the EPR results described in the following correspond to Fe(II)/H2O2 titrations. The results of the spin quantification are summarized in Table 1 for all the titrations reported in this investigation.
X-band CW-EPR spectra of Listeria innocua Dps at different stages of iron oxidation: a by O2 under aerobic condition and b by H2O2 under anaerobic conditions, starting from an apoprotein containing 1.5 Fe(III)/subunit. Experimental conditions are reported in Materials and methods (color figure online)
Figure 3 shows the effect of adding different amounts of Fe (II)/H2O2 to the apoprotein in detail. Six additions of Fe (II) up to 120 Fe(II)/monomer were carried out. In the titration, the signal intensity reaches a maximum upon addition of 12 Fe (II)/monomer, while successive additions produce a signal reduction. The maximum signal intensity accounts for ~ 11.5 Fe(III)/dodecamer as estimated using the apoprotein as the intensity standard (see Table 1). The numerous Fe(II)/H2O2 titrations of Listeria Dps, under many different experimental conditions including different protein concentrations (see Fig. S2), have clearly shown a definite trend: a linear increase of the EPR signal relative to the formation of mononuclear Fe(III) reaching the maximum between the 12/1 and 24/1 iron/protein ratio. Complete disappearance of the EPR signal occurs only in the presence of the iron core (~ 300 iron/protein) as shown in Fig. S3. While no significant variation of the spectral shape is produced during the titration (see Fig. S4), the contribution of different features to the EPR line varies after 2 days storage of the sample at 4 °C (see Fig. S5). The signal lineshape is not isotropic and therefore points to the presence of multiple g′ = 4.3 Fe (III) sites and migration occurs in time among sites.
In the present study, the site-specific mutants of the Listeria Dps ferroxidase center were also characterized. Both the A and the B sites contain a histidine ligand, namely, His31 and His43. To establish their contribution to the functionality of the ferroxidase center, these histidine residues were replaced by glycine in the H31G and H43G mutants. Asp58, which is an additional A site iron ligand, was substituted by alanine in the triple mutant, H31G-H43G-D58A. Titration experiments were performed also on the mutants. In the triple mutant of Dps, the amount of detected Fe(III) is always negligible despite the level of Fe(II) addition. In Fig. 4, the EPR signal corresponding to a 12 Fe(II)/H2O2 addition is compared for the native Dps and the triple variant. This result clearly demonstrate that the EPR signal detected in the wt Listeria Dps is specific for the iron bound to the ferroxidase centers, since the corresponding signal is missing in the H31G-H43G-D58A mutant.
EPR titrations performed on the H31G and H43G mutants give further important information, as shown in Fig. 5. The EPR titration performed on the H31G mutant reaches approximately the same amount of mononuclear Fe(III) observed in the native Dps protein, namely a maximum of ~ 9.0 mononuclear Fe(III)/dodecamer. Spectral analysis of the EPR signal for the H31G mutant shows that while at different stages of the Fe(II)/H2O2 titration the spectral shape is conserved, comparison with the corresponding spectrum of native Dps after 12 Fe(II) addition highlights a broader shape for the signal at g = 4.3 (see Fig. S6), indicative of a different environment for the mononuclear Fe(III) species. Conversely, in the H43G mutant we detect a signal for mononuclear Fe(III) corresponding to less than 1.0 Fe(III)/subunit and no significant variations of the signal intensity during titration.
As already discussed, the investigation was carried out both on native Dps in the apo form and in the presence of the iron core (see Fig. S3). Also, all the site-directed mutants were provided with the mineralization core. The EPR spectra were recorded on the proteins with the iron core to check if the signal intensity could account for the lack of iron found in the ferroxidase center by X-ray crystallography on Dps crystals prepared in the same conditions. The apoprotein shows a larger amount of Fe(III)/monomer than the protein in the presence of the iron core. In all mutants, an insignificant amount of mononuclear Fe(III) has been detected, i.e. < 1 Fe(III)/monomer. Data relative to the triple mutants are not shown, because the signal intensity is even weaker. The disappearance of the EPR signal in the presence of large excess of iron cannot be explained in terms of the formation of anti-ferromagnetically coupled Fe(III)–Fe(III) dimers since in the mutant dimers cannot be formed, due to the lack of the iron ligand His31 or His43. The EPR signal of the mononuclear Fe(III), located in the ferroxidase center, is not detectable in the presence of the iron core due to the magnetic relaxation effect produced by the presence of a superparamagnetic cluster or to the migration of iron from the ferroxidase center into the internal cavity. X-ray data seem to confirm this last hypothesis, since the Dps from E. coli loaded with 300 iron/ dodecamer does not contain iron ions at the ferroxidase center [24].
Finally, EPR experiments were performed also on the wt Dps from Listeria monocytogenes as shown in Fig. 6. The titration is characterized by an analogous trend as for Dps from Listeria innocua, proving that the mechanism of iron oxidation and incorporation coupled to the hydrogen peroxide detoxification takes place in a similar manner in all the members of the Dps family.
4 Discussion
The ferroxidase centers in Dps proteins are unusual because they are not placed, as in the canonical ferritins, in the middle of the four-helix bundle but at the interface of the two symmetry-related subunits. Thus, the basic structural and functional units of Dps are the dimers, which are more stable than in the other members of the ferritin superfamily [25]. As a consequence, the ferroxidation reaction has distinctive features compared to canonical ferritins.
The present EPR studies on the wt Listeria Dps, where the ferroxidase center was first described and characterized, and on three site-specific ferroxidase center mutants provide insight into the role played by specific iron ligands and by the ferroxidase center itself in the iron oxidation/uptake and in hydrogen peroxide detoxification process.
Previous studies underlined that ferroxidase activity requires binding of two Fe(II) to the A and B sites of the catalytic center and it depends on the multiplicity of iron–protein interactions thereby established (see Fig. 7 left panel). In canonical ferritins, only site A harbors a histidine residue, whereas in Listeria Dps the histidine residues are present in both A and B sites (His31 and His43 respectively). The X-ray crystal structure of Dps disclosed the presence of only one iron ion per monomer at the ferroxidase center bound to the A site and coordinated by a His31 Asp58, Glu62, and a water molecule which in turn is hydrogen bonded to His43, indicating that probably A has a higher affinity for iron with respect to B.
Iron binding sites at the ferroxidase center of Listeria innocua Dps. Left panel: structural representation of the A, B and C sites at the ferroxidase center of the Listeria innocua Dps (Pdb code:1QHG). The B and C sites are modeled on the basis of the position of water molecules bound to the center in the wt protein (PDB code:1QHG) and H43G mutant (PDB code: 2BKC), respectively. Alternative conformation of Asp58 and Glu62 possibly coordinating iron in the B and C positions have been modeled manually starting from the most probable conformers of the two residues. Middle panel: possible position of the iron ion in the ferroxidase center upon mutation of the His 31 into Gly. The iron ion was directly modeled in the ferroxidase center of the H31G mutant (PDB code: 2BK6). Right panel: superimposition between the wt Dps from Listeria innocua (in gray) and the H43G mutant (PDB code: 2BKC) where the twofold symmetry-related monomer are colored in salmon and blue. The iron ion of the A site is indicated as orange sphere. The pictures were generated using PyMOL (The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC).
Previous studies have already demonstrated that differently from the ferritin, the hydrogen peroxide is a better oxidant for iron than oxygen. The EPR studies presented here confirm this hypothesis; indeed, the iron binding and oxidation at the ferroxidase center take place with both oxygen and hydrogen peroxide. To gain insight into the iron oxidation and incorporation process coupled with hydrogen peroxide reduction, the EPR signal was measured upon addition to the protein sample of Fe (II) and hydrogen peroxide on the basis of the following reaction:
The maximum of the signal at g = 4.3 is reached when 12 Fe(II) are added to the protein sample, indicating that the iron oxidation takes place probably with the saturation of iron in site A. The EPR data collected on the triple mutant (H31G, H43G, D58A) show that in the same condition, there is no significant signal at g = 4.3, indicating that the binding of Fe(II) and the subsequent iron oxidation take place specifically at the ferroxidase center. The EPR signal measured upon addition of Fe(II) and H2O2 to the H31G mutant indicates that iron is still able to bind to the iron center (even if with lower affinity), but the spectral shape changes indicate that Fe(III) binds to different residues. The X-ray structure of the H31G mutant (PDB code 2BK6) may provide a possible explanation of this phenomenon. Indeed, the mutation of His31 induces small changes in the center, and, as shown in the middle panel of Fig. 7, iron (modeled starting from a water molecule bound to the center of H31G mutant) may be bound in a position shifted with respect to that of site A, possibly coordinated by D58 and E62 and by Asp 47.
The EPR spectra collected on the H43G mutant, in the same experimental condition, dramatically change the EPR signal that becomes very small. On the other hand, the X-ray structure of the mutant H43G, solved by Ilari et al. [12], shows that the mutation does not change the geometry of the center (Fig. 7, right panel), and the fluorescence titration presented in the same paper, carried out in the presence of hydrogen peroxide, shows that the center, upon H43G mutation, is still able to bind two iron ions. Altogether, these data and the EPR titration suggest that the iron may bind to site A and to another site named site C, modeled in the left panel of Fig. 7 on the basis of the H43G mutant coordinates (PDB code: 2BKC). We hypothesized that no EPR signal is detected, since the distance between the two sites is about 3 Å allowing the Fe(III)–Fe(III) spin coupling with the consequent silencing of the signal at g = 4.3.
Moreover, since these sites are both occupied during the titration in the H43 mutant, the data seem to suggest a cooperative binding, i.e., the binding of iron to site A may facilitate the binding of iron to site C in the absence of H43. Indeed, as shown in the left panel of Fig. 7 the binding of iron to the A site could cause the displacement of E62 and facilitate the rotation of D58, which can acquire the right position to bind iron in the C site.
Finally, the EPR titrations show that, when the first Fe(II) is added to Listeria Dps in the presence of H2O2, the EPR signal at g = 4.3 increases proving that iron is oxidized at the ferroxidase center occupying site A, previously identified by X-ray crystallography. When the addition of iron exceeds 12/dodecamer in the presence of H2O2, the EPR signal starts to decrease and iron starts occupying the B site placed at 3 Å from the A site (as hypothesized by Ilari et al. [3]).
The EPR signal disappears when ~ 300 iron/dodecamer are added, indicating the migration of Fe(III) to the internal cavity with the formation of an antiferromagnetic oxyhydroxide iron core [26].
5 Conclusions
The data presented in this paper confirm the importance of the H31, H43 and D58 residues for the iron oxidation/H2O2 reduction reaction. These residues allow the binding of iron to the ferroxidase center and its oxidation; their mutation causes the loss of the Dps detoxification activity (triple mutant) or the modification of the iron binding and oxidation mode (H31G and H43G mutants). Moreover, the EPR data suggest that the ferroxidase center is endowed with a unique flexibility, allowing the iron binding and its oxidation to the center even if one of the two histidines is mutated; this plasticity is necessary for the migration of iron to the negatively charged inner cavity.
The results presented in this paper complete the data acquired on the last 20 years on the detoxification mechanism catalyzed by Dps proteins in bacteria and allow us to finally hypothesize a complete mechanism for iron oxidation and incorporation. On the basis of the present results, we can conclude that the mechanism of iron oxidation by hydrogen peroxide could take place also if only the A site is occupied by iron by two steps summarized as follows:
-
1.
\({\text{Fe}}\left( {{\text{II}}} \right) \, \left( {\text{site A}} \right) + {\text{H}}_{2} {\text{O}}_{2} + 2{\text{H}}^{ + } \to {\text{Fe}}\left( {{\text{III}}} \right)\left( {\text{site A}} \right) + {\text{radical}} + 2{\text{H}}_{2} {\text{O}}{.}\)
-
2.
\({\text{Radical}} + {\text{Fe}}\left( {{\text{II}}} \right)\left( {\text{site B}} \right) \to {\text{Fe}}\left( {{\text{III}}} \right)\left( {\text{site B}} \right)\)
The first reaction which takes place in the ferroxidase center is the binding of Fe(II) to the A site as previously demonstrated by anaerobic fluorimetric titrations [12]. Then, iron is oxidized to Fe(III) in the site A of the ferroxidase center as demonstrated by EPR experiments with the formation of a tryptophan radical, as visualized by Bellaparona et al. with kinetic experiments [27]. A second iron atom is then oxidized at the B site and, as a consequence of the spin coupling, the EPR signal decreases. Finally, the iron ion leaves the A and B sites, hopping to other negatively charged amino acids that form the iron core nucleation site, namely Glu 44 and Asp 47, facing toward the inner cavity where the mineral oxyhydroxide iron core forms [24].
In conclusion, all the data point to the fact that in Dps the oxidation of two iron ions at the ferroxidase center by a hydrogen peroxide molecule takes place sequentially with the formation of a transient tryptophan radical, whereas the migration of iron to the inner cavity is preceded by the formation of a Fe(III)–Fe(III) dimer as suggested by EPR titrations.
References
M. Almiron, A.J. Link, F. Deirdre, R. Kolter, Genes Dev. 6, 2646–2654 (1992)
M. Bozzi, G. Mignogna, S. Stefanini, D. Barra, C. Longhi, P. Valenti, E. Chiancone, J. Biol. Chem. 272, 3259–3265 (1997)
A. Ilari, S. Stefanini, E. Chiancone, D. Tsernoglou, Nat. Struct. Biol. 7, 38–43 (2000)
R.A. Grant, D.J. Filman, S.E. Finkel, R. Kolter, J.M. Hogle, Nat. Struct. Biol. 5, 294–303 (1998)
V.O. Karas, I. Westerlaken, A.S. Meyer, J. Bacteriol. 197, 3206–3215 (2015)
G. Zhao, P. Ceci, A. Ilari, L. Giangiacomo, T.M. Laue, E. Chiancone, N.D. Chasteen, J. Biol. Chem. 277, 27689–27696 (2002)
G. Zhao, F. Bou-Abdallah, P. Arosio, S. Levi, C. Janus-Chandler, N.D. Chasteen, Biochemistry 42, 3142–3150 (2003)
J. Bunker, T. Lowry, G. Davis, B. Zhang, D. Brosnahan, S. Lindsay, R. Costen, S. Choi, P. Arosio, G.D. Watt, Biophys. Chem. 114, 235–244 (2005)
S. Franceschini, P. Ceci, F. Alaleona, E. Chiancone, A. Ilari, FEBS J. 273, 4913–4928 (2006)
P. Ceci, L. Mangiarotti, C. Rivetti, E. Chiancone, Nucleic Acids Res. 35, 2247–2256 (2007)
F. Tonello, W.G. Dundon, B. Satin, M. Molinari, G. Tognon, G. Grandi, G. Del Giudice, R. Rappuoli, C. Montecucco, Mol. Microbiol. 34, 238–246 (1999)
A. Ilari, M.C. Latella, P. Ceci, F. Ribacchi, M. Su, L. Giangiacomo, S. Stefanini, N.D. Chasteen, E. Chiancone, Biochemistry 44, 5579–5587 (2005)
M. Polidoro, D. De Biase, B. Montagnini, L. Guarrera, S. Cavallo, P. Valenti, S. Stefanini, E. Chiancone, Gene 296, 121–128 (2002)
A. Genoni, G. La Ganga, A. Volpe, F. Puntoriero, M. Di Valentin, M. Bonchio, M. Natali, A. Sartorel, Faraday Discuss. 185, 121–141 (2015)
A. Zoleo, M. Brustolon, A. Barbon, A. Silvestri, G. Molin, S. Tonietto, J. Cult. Herit. 16, 322–328 (2015)
M.R. Cheesman, N.E. le Brun, F.H.A. Kadir, A.J. Thomson, G.R. Moore, S.C. Andrews, J.R. Guest, P.M. Harrison, J.M.A. Smith, S.J. Yewdall, Biochem. J. 292, 47–56 (1993)
V. Preger, N. Tango, C. Marchand, S.D. Lemaire, D. Carbonera, M. Di Valentin, A. Costa, P. Pupillo, P. Trostet, Plant Physiol. 150, 606–620 (2009)
J.M. Bradley, D.A. Svistunenko, J. Pullin, N. Hill, R.K. Stuart, B. Palenik, M.T. Wilson, A.M. Hemmings, G.R. Moore, N.E. Le-Brun, PNAS 116, 2058–2067 (2019)
J.M. Bradley, J. Pullin, G.R. Moore, D.A. Svistunenko, A.M. Hemmings, N.E. Le Brun, Dalton Trans. 49, 1545–1554 (2020)
F. Bou-Abdallah, H. Yang, A. Awomolo, B. Cooper, M.R. Woodhall, S.C. Andrews, N.D. Chasteen, Biochemistry 53, 483–495 (2014)
J.M. Bradley, D.A. Svistunenko, G.R. Moore, N.E. Le Brun, Metallomics 9, 1421–1428 (2017)
E.I. Solomon, T.C. Brunold, M.I. Davis, J.N. Kemsley, S.K. Lee, N. Lehnert, F. Neese, A.J. Skulan, Y.S. Yang, J. Zhou, Chem. Rev. 100, 235–349 (2000)
F. Bou-Abdallah, N.D. Chasteen, J. Biol. Inorg. Chem. 13, 15–24 (2008)
A. Ilari, P. Ceci, D. Ferrari, G.L. Rossi, E. Chiancone, J. Biol. Chem. 277, 37619–37623 (2002)
R. Chiaraluce, V. Consalvi, S. Cavallo, A. Ilari, S. Stefanini, E. Chiancone, Eur. J. Biochem. 267, 5733–5741 (2000)
Y. Gossuin, R.N. Muller, P. Gillis, NMR Biomed. 17, 427–432 (2004)
G. Bellapadrona, M. Ardini, P. Ceci, S. Stefanini, E. Chiancone, Free Radic. Biol. Med. 48, 292–297 (2010)
Acknowledgements
This paper is dedicated to Emilia Chiancone and Giovanni Giacometti who passed away recently. They started together this project with enthusiasm and inspired the content of this work.
Funding
Open access funding provided by Università degli Studi di Padova within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ilari, A., Bellapadrona, G., Carbonera, D. et al. Disclosing the Molecular Mechanism of Iron Incorporation in Listeria innocua Dps by EPR Spectroscopy. Appl Magn Reson 51, 1543–1557 (2020). https://doi.org/10.1007/s00723-020-01287-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00723-020-01287-x