Abstract
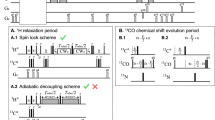
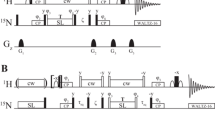
Characterization of protein solutions is of great importance for biophysical research, pharmaceutical industry, and medicine. Particularly, the monitoring of the protein aggregation is crucial at all stages of biotechnological production and in the diagnosis of dangerous diseases. The present work is focused on a study of prospects and possibilities of NMR relaxation of solvent nuclei for monitoring the state of proteins in solutions. The spin–lattice and spin–spin relaxation rates (R1 and R2) of solvent nuclei were measured in the solutions of a small globular protein, RRM2 domain of TDP-43 protein. The solvent was either H2O- or D2O-based buffer with pH 6.5 and contained 20 mM sodium phosphate and 150 mM NaCl. The relaxation rates of the solvent 1H, 2H, 23Na, and 35Cl nuclei in solutions of soluble and aggregated RRM2 domain of TDP-43 protein were studied. The aggregation was induced by mild oxidative stress, using treatment by hydrogen peroxide. It was found that aggregation of protein could be detected using NMR relaxation of 1H nuclei. The observed CPMG dispersion for R2 rates confirms the millisecond timescale for the hydrogen exchange between water and protein sites. The correlation times and binding constants for sodium and chlorine ions were estimated using concentration dependences for relaxation rates (23Na, 35Cl). The relaxation rates of solvent nuclei are sensitive to the presence of protein in solution even at low protein concentrations, and the relaxation rates of different nuclei reflect various aspects of the state of the protein.





Similar content being viewed by others
References
C. Haass, D.J. Selkoe, Nat. Rev. Mol. Cell Biol. 8, 101 (2007)
T. Strohäker, B.C. Jung, S.-H. Liou, C.O. Fernandez, D. Riedel, S. Becker, G.M. Halliday, M. Bennati, W.S. Kim, S.-J. Lee, M. Zweckstetter, Nat. Commun. 10, 5535 (2019)
B. Nizynski, W. Dzwolak, K. Nieznanski, Protein Sci. 26, 2126 (2017)
A.S. Chen-Plotkin, V.M.-Y. Lee, J.Q. Trojanowski, Nat. Rev. Neurol. 6, 211 (2010)
S.K. Maji, M.H. Perrin, M.R. Sawaya, S. Jessberger, K. Vadodaria, R.A. Rissman, P.S. Singru, K.P.R. Nilsson, R. Simon, D. Schubert, D. Eisenberg, J. Rivier, P. Sawchenko, W. Vale, R. Riek, Science 325, 328 (2009)
P.S. Dannies, Mol. Genet. Metab. 76, 6 (2002)
W.K. Schmidt, H.P. Moore, J. Biol. Chem. 269, 27115 (1994)
Y. Li, C.J. Roberts, in Aggregation of Therapeutic Proteins, ed. by W. Wang, C.J. Roberts (Wiley, Hoboken, 2010), pp. 63–102
S. Gregoire, J. Irwin, I. Kwon, Korean J. Chem. Eng. 29, 693 (2012)
Z. Yu, J.C. Reid, Y.-P. Yang, J. Pharm. Sci. 102, 4284 (2013)
D.L. Melnikova, V.D. Skirda, I.V. Nesmelova, J. Phys. Chem. B 121, 2980 (2017)
I.V. Nesmelova, D.L. Melnikova, V. Ranjan, V.D. Skirda, Progr. Mol. Biol. Transl. Sci. 166, 85 (2019)
S. Kiihne, R.G. Bryant, Biophys. J. 78, 2163 (2000)
B.P. Hills, S.F. Takacs, P.S. Belton, Mol. Phys. 67, 903 (1989)
B.P. Hills, S.F. Takacs, P.S. Belton, Mol. Phys. 67, 919 (1989)
M.B. Taraban, R.A. DePaz, B. Lobo, Y.B. Yu, Anal. Chem. 89, 5494 (2017)
B. Halle, Philos. Trans. R. Soc. B Biol. Sci. 359, 1207 (2004)
V.P. Denisov, B. Halle, J. Am. Chem. Soc. 124, 10264 (2002)
J.S. Leigh, J. Magn. Reson. 1969(4), 308 (1971)
S.O. Rabdano, A.V. Donets, M.A. Vovk, D. Michel, V.I. Chizhik, J. Phys. Chem. B 119, 13358 (2015)
M.B. Taraban, R.A. DePaz, B. Lobo, Y.B. Yu, Anal. Chem. 91, 4107 (2019)
J.P. Carver, R.E. Richards, J. Magn. Reson. 1969(6), 89 (1972)
K.A. Valiev, J. Struct. Chem. 3, 630 (1962)
A. Geiger, H.G. Hertz, Adv. Mol. Relax. Process. 9, 293 (1977)
V.I. Chizhik, Y.S. Chernyshev, A.V. Donets, V.V. Frolov, A.V. Komolkin, M.G. Shelyapina, in Magnetic Resonance and Its Applications (Springer, Cham, 2014)
T.L. James, J.H. Noggle, Proc. Natl. Acad. Sci. USA 62, 644 (1969)
A.M. Torres, D.J. Philp, R. Kemp-Harper, C. Garvey, P.W. Kuchel, Magn. Reson. Chem. 43, 217 (2005)
T. Janc, M. Lukšič, V. Vlachy, B. Rigaud, A.-L. Rollet, J.-P. Korb, G. Mériguet, N. Malikova, Phys. Chem. Chem. Phys. 20, 30340 (2018)
T. Minami, W.S. Price, D.J. Cutler, J. Pharm. Sci. 81, 419 (1992)
W.S. Price, P.W. Kuchel, B.A. Cornell, Biophys. Chem. 40, 329 (1991)
W.S. Price, N.H. Ge, L.Z. Hong, L.P. Hwang, J. Am. Chem. Soc. 115, 1095 (1993)
R.L. Ward, Arch. Biochem. Biophys. 169, 22 (1975)
S.O. Rabdano, S.A. Izmailov, D.A. Luzik, A. Groves, I.S. Podkorytov, N.R. Skrynnikov, Sci. Rep. 7, 11161 (2017)
D. Wierzuchowska, L.W. Skórski, B. Blicharska, Acta Phys. Pol. A 129, 226 (2016)
A. Jerschow, N. Müller, J. Magn. Reson. 125, 372 (1997)
J.J. Helmus, C.P. Jaroniec, J. Biomol. NMR 55, 355 (2013)
SciPy 1.0 Contributors, P. Virtanen, R. Gommers, T.E. Oliphant, M. Haberland, T. Reddy, D. Cournapeau, E. Burovski, P. Peterson, W. Weckesser, J. Bright, S.J. van der Walt, M. Brett, J. Wilson, K.J. Millman, N. Mayorov, A.R.J. Nelson, E. Jones, R. Kern, E. Larson, C.J. Carey, İ. Polat, Y. Feng, E.W. Moore, J. VanderPlas, D. Laxalde, J. Perktold, R. Cimrman, I. Henriksen, E.A. Quintero, C.R. Harris, A.M. Archibald, A.H. Ribeiro, F. Pedregosa, P. van Mulbregt, Nat. Methods 17, 261 (2020)
V.V. Krishnan, J. Magn. Reson. 124, 468 (1997)
D. Wierzuchowska, M. Witek, B. Blicharska, Acta Phys. Pol. A 137, 21 (2020)
R.R. Knispel, M.M. Pintar, Chem. Phys. Lett. 32, 238 (1975)
M. Pfuhl, HoA Chen, SørenM Kristensen, PaulC Driscoll, J. Biomol. NMR 14, 307 (1999)
V. Levi, F.L. González Flecha, Biochim. Biophys. Acta BBA Proteins Proteom. 1599, 141 (2002)
Z. Liu, W.-P. Zhang, Q. Xing, X. Ren, M. Liu, C. Tang, Angew. Chem. Int. Ed. 51, 469 (2012)
M.E.M. Cromwell, E. Hilario, F. Jacobson, AAPS J. 8, E572 (2006)
N.-H. Ge, W.S. Price, L.-Z. Hong, L.-P. Hwang, J. Magn. Reson. 1969(97), 656 (1992)
E. Canet, D. Mammoli, P. Kadeřávek, P. Pelupessy, G. Bodenhausen, Phys. Chem. Chem. Phys. 18, 10144 (2016)
S. Scheiner, M. Čuma, J. Am. Chem. Soc. 118, 1511 (1996)
A. Donets, V. Chizhik, Struct. Chem. 22, 465 (2011)
V.I. Chizhik, Mol. Phys. 90, 653 (1997)
M.M. Civan, M. Shporer, in Biological Magnetic Resonance, ed. by L.J. Berliner, J. Reuben (Springer, Boston, 1978), pp. 1–32
V.I. Chizhik, I.S. Podkorytov, A.P. Kaikkonen, V.I. Mikhailov, J. Magn. Reson. A 123, 1 (1996)
B. Hoffmann, C. Eichmüller, O. Steinhauser, R. Konrat, Methods Enzymol. 394, 142–175 (2005)
A. Schedlbauer, N. Coudevylle, R. Auer, K. Kloiber, M. Tollinger, R. Konrat, J. Am. Chem. Soc. 131, 6038 (2009)
A.C. Liwang, A. Bax, J. Magn. Reson. 127, 54 (1997)
R. Eggenberger, S. Gerber, H. Huber, D. Searles, M. Welker, J. Chem. Phys. 97, 5898 (1992)
V.I. Chizhik, A.V. Egorov, M.S. Pavlova, M.I. Egorova, A.V. Donets, J. Mol. Liq. 224, 730 (2016)
Acknowledgements
We would like to thank Prof. Nikolai Skrynnikov for the initial discussions about the idea of the study of protein solutions and aggregation problem using NMR relaxation of water nuclei; Mikhail Vovk and Vladislav Salikov for the help with collection and processing 2H, 23Na, and 35Cl relaxation data; Dr. Boris Kharkov for the help with a collection of 1H diffusion data. The research was supported by RFBR and CITMA according to project #18-53-34003. Most of NMR measurements were performed at the Research park of St. Petersburg State University “Center for Magnetic Resonance”.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Rabdano, S.O., Bystrov, S.S., Luzik, D.A. et al. NMR Relaxation of Nuclei of Buffer as a Probe for Monitoring Protein Solutions Including Aggregation Processes. Appl Magn Reson 51, 1653–1668 (2020). https://doi.org/10.1007/s00723-020-01227-9
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00723-020-01227-9