Abstract
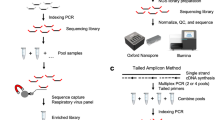
Surveillance of the SARS-CoV-2 genome has become a crucial technique in the management of COVID-19, aiding the pandemic response and supporting effective public health interventions. Typically, whole-genomic sequencing is used along with PCR-based target enrichment techniques to identify SARS-CoV-2 variants, which is a complicated and time-consuming process that requires central laboratory facilities. Thus, there is an urgent need to develop rapid and cost-effective tools for precise on-site detection and identification of SARS-CoV-2 strains. In this study, we demonstrate the rapid diagnosis of COVID-19 and identification of SARS-CoV-2 variants by amplification and sequencing of the entire SARS-CoV-2 S gene using isothermal enzymatic recombinase amplification combined with the advanced Oxford nanopore sequencing technique. The entire procedure, from sampling to sequencing, takes less than 8 hours and can be performed with limited resources. The newly developed method has noteworthy implications for examining the transmission dynamics of the virus, detecting novel genetic variants, and assessing the effect of mutations on diagnostic approaches, antiviral treatments, and vaccines.







Similar content being viewed by others
Data availability
The data supporting the findings of this study are presented in the article and its supplementary materials and are available from the corresponding author upon request.
References
Choi JY, Smith DM (2021) SARS-CoV-2 Variants of Concern. Yonsei Med J 62:961–968. https://doi.org/10.3349/ymj.2021.62.11.961
Saravanan KA, Panigrahi M, Kumar H et al (2022) Role of genomics in combating COVID-19 pandemic. Gene 823:146387. https://doi.org/10.1016/j.gene.2022.146387
Barbé L, Schaeffer J, Besnard A et al (2022) SARS-CoV-2 Whole-Genome Sequencing Using Oxford Nanopore Technology for Variant Monitoring in Wastewaters. Front Microbiol 13:889811. https://doi.org/10.3389/fmicb.2022.889811
Chiara M, D’Erchia AM, Gissi C et al (2020) Next generation sequencing of SARS-CoV-2 genomes: challenges, applications and opportunities. https://doi.org/10.1093/bib/bbaa297. Brief Bioinform bbaa297
Xiao M, Liu X, Ji J et al (2020) Multiple approaches for massively parallel sequencing of SARS-CoV-2 genomes directly from clinical samples. Genome Med 12:57. https://doi.org/10.1186/s13073-020-00751-4
Lin B, Hui J, Mao H (2021) Nanopore Technology and Its Applications in Gene Sequencing. Biosens (Basel) 11:214. https://doi.org/10.3390/bios11070214
MacKenzie M, Argyropoulos C (2023) An Introduction to Nanopore Sequencing: Past, Present, and Future Considerations. Micromachines 14:459. https://doi.org/10.3390/mi14020459
Barbé L, Schaeffer J, Besnard A et al (2022) SARS-CoV-2 Whole-Genome Sequencing Using Oxford Nanopore Technology for Variant Monitoring in Wastewaters. Front Microbiol 13. https://doi.org/10.3389/fmicb.2022.889811
Kozarewa I, Armisen J, Gardner A et al (2015) Overview of Target Enrichment Strategies. Current protocols in molecular biology / edited by Frederick M Ausubel. 112:7.21.1–7.21.23. https://doi.org/10.1002/0471142727.mb0721s112
Foo PC, Nurul Najian AB, Muhamad NA et al (2020) Loop-mediated isothermal amplification (LAMP) reaction as viable PCR substitute for diagnostic applications: a comparative analysis study of LAMP, conventional PCR, nested PCR (nPCR) and real-time PCR (qPCR) based on Entamoeba histolytica DNA derived from faecal sample. BMC Biotechnol 20:34. https://doi.org/10.1186/s12896-020-00629-8
Stüder F, Petit J-L, Engelen S, Mendoza-Parra MA (2021) Real-time SARS-CoV-2 diagnostic and variants tracking over multiple candidates using nanopore DNA sequencing. Sci Rep 11:15869. https://doi.org/10.1038/s41598-021-95563-w
Alhamid G, Tombuloglu H, Rabaan AA, Al-Suhaimi E (2022) SARS-CoV-2 detection methods: A comprehensive review. Saudi J Biol Sci 29:103465. https://doi.org/10.1016/j.sjbs.2022.103465
Recordon-Pinson P, Blondot M-L, Bellecave P et al (2021) A Simple and Fast Method to Sequence the Full-Length Spike Gene for SARS-CoV-2 Variant Identification from Patient Samples. COVID 1:337–344. https://doi.org/10.3390/covid1010028
Jackson CB, Farzan M, Chen B, Choe H (2022) Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol 23:3–20. https://doi.org/10.1038/s41580-021-00418-x
Xia S, Chen X (2020) Ultrasensitive and Whole-Course Encapsulated Field Detection of 2019-nCoV Gene Applying Exponential Amplification from RNA Combined with Chemical Probes. https://doi.org/10.26434/chemrxiv.12012789.v1
Xia S, Chen X (2020) Single-copy sensitive, field-deployable, and simultaneous dual-gene detection of SARS-CoV-2 RNA via modified RT–RPA. Cell Discov 6:1–4. https://doi.org/10.1038/s41421-020-0175-x
Cuong HQ, Hai ND, Linh HT et al (2021) The Production of Standardized Samples with Known Concentrations for Severe Acute Respiratory Syndrome Coronavirus 2 RT-qPCR Testing Validation for Developing Countries in the Period of the Pandemic Era. Biomed Res Int 2021:5516344. https://doi.org/10.1155/2021/551634
Tran DH, Cuong HQ, Tran HT et al (2021) A comparative study of isothermal nucleic acid amplification methods for SARS-CoV-2 detection at point-of-care. Chem Biology Lett 8:106–116
Wilrich C, Wilrich P-T (2009) Estimation of the POD function and the LOD of a qualitative microbiological measurement method. J AOAC Int 92:1763–1772
Burrell CJ, Howard CR, Murphy FA (2017) Laboratory Diagnosis of Virus Diseases. Fenner White’s Med Virol 135–154. https://doi.org/10.1016/B978-0-12-375156-0.00010-2
Ranasinghe D, Jayadas TTP, Jayathilaka D et al (2022) Comparison of different sequencing techniques for identification of SARS-CoV-2 variants of concern with multiplex real-time PCR. PLoS ONE 17:e0265220. https://doi.org/10.1371/journal.pone.0265220
Lu H, Giordano F, Ning Z (2016) Oxford Nanopore MinION Sequencing and Genome Assembly. Genomics. Proteom Bioinf 14:265–279. https://doi.org/10.1016/j.gpb.2016.05.004
(2022) The Midnight Kit: sequencing whole SARS-CoV-2 genomes with Oxford Nanopore. In: Oxford Nanopore Technologies. https://nanoporetech.com/knowledge-exchange/on-demand/sequencing-whole-SARS-CoV-2-genomes. Accessed 1 Dec 2023
von Koskela A, Lindqvist CM, Asghar N et al (2023) Comparison of SARS-CoV-2 whole genome sequencing using tiled amplicon enrichment and bait hybridization. Sci Rep 13:6461. https://doi.org/10.1038/s41598-023-33168-1
Bull RA, Adikari TN, Ferguson JM et al (2020) Analytical validity of nanopore sequencing for rapid SARS-CoV-2 genome analysis. Nat Commun 11:6272. https://doi.org/10.1038/s41467-020-20075-6
Author information
Authors and Affiliations
Contributions
HTTP and MTV led the conceptualization and design of the study, formulated the research questions, and supervised the laboratory work. DHT conducted the major experimental procedures and collected the data. HDKD and HDN performed sequencing data analysis and interpretation. HTT and TNMP provided technical support in the laboratory and assisted in data interpretation. HTL and HQC provided the samples, assisted in experimental design, and contributed to manuscript editing. DHT, HDKD, MTV, and HTTP contributed to manuscript writing, editing, and critical revisions. All authors contributed to the intellectual content and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethical approval
The study was approved by the Research Ethics Committee of Pasteur Institute in Ho Chi Minh City, Vietnam.
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Communicated by Pablo Pineyro
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tran, D.H., Do, H.D.K., Tran, H.T. et al. Rapid identification of SARS-CoV-2 strains via isothermal enzymatic recombinase amplification and nanopore sequencing. Arch Virol 169, 87 (2024). https://doi.org/10.1007/s00705-024-06012-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00705-024-06012-8