Abstract
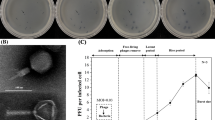
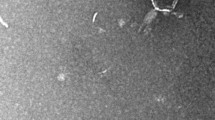
Vibrio parahaemolyticus is a widely recognized pathogen that has caused numerous outbreaks and is prevalent in the marine environment. In this study, we investigated the characteristics of the novel V. parahaemolyticus strain BTXS2 and its associated phage, VB_VpP_BT-1011, isolated from the Bohai Coast (Tianjin, China). Strain BTXS2 is a short coryneform bacterium with a terminal flagellum and is able to utilize and metabolize a wide variety of organic matter because of its unique carbon source utilization and enzyme activity. It grows well in medium between pH 5.0 and 9.0 and salinities of simulated freshwater, estuary water, and seawater (NaCl 0.5%-3%). Multiple antibiotic resistance genes and virulence genes that endanger human health were found in the BTXS2 genome. Phage VB_VpP_BT-1011, which infects BTXS2, is a 40,065-bp double-stranded DNA virus of the family Myoviridae with a latent time of 30 min and burst size of 24 PFU/cell. Like its host, the phage tolerates a broad range of environmental conditions (salinity, 0-3% NaCl; pH 5.0-9.0; temperature, 4-37°C). A host range test showed that the phage only infected and inhibited isolate BTXS2. In summary, we investigated a novel V. parahaemolyticus host-phage pair and the antibacterial effect of the phage on V. parahaemolyticus, providing insights into marine microbial ecology and risks.








Similar content being viewed by others
Availability of data and materials
The nucleotide sequence of V. parahaemolyticus BTXS2 can be viewed in the NCBI database (https://www.ncbi.nlm.nih.gov/nuccore/?term=BTXS2), and the nucleotide sequence of the phage can be viewed at https://www.ncbi.nlm.nih.gov/nuccore/MW009675.1.
References
Wang R, Zhong Y, Gu X, Yuan J, Saeed AF, Wang S (2015) The pathogenesis, detection, and prevention of Vibrio parahaemolyticus. Front Microbiol 6:144
Baker-Austin C, Oliver JD, Alam M, Ali A, Waldor MK, Qadri F, Martinez-Urtaza J (2018) Vibrio spp. infections. Nat Rev Dis Primers 4:8
Wang Y, Shen X-S, Gu RR, Shi YF, Tian LL (2015) Application of a rapid method for detecting Vibrio parahaemolyticus in seafood. J Food Saf 35:26–31
Fujino T, Okuno Y, Nakada D, Aoyama A, Ueho T (1953) On the bacteriological examination of Shirasu food poisoning. Medjosaka Univ 4:299–304
Quilici ML, Robert-Pillot A, Picart J, Fournier JM (2005) Pandernic Vibrio parahaemolyticus O3: K6 spread, France. Emerg Infect Dis 11:1148–1149
Martinez-Urtaza J, Simental L, Velasco D, DePaola A, Ishibashi M, Nakaguchi Y, Nishibuchi M, Carrera-Flores D, Rey-Alvarez C, Pousa A (2005) Pandemic Vibrio parahalemolyticus O3: K6, Europe. Emerg Infect Dis 11:1319–1320
Martinez-Urtaza J, Trinanes J, Abanto M, Lozano-Leon A, Llovo-Taboada J, Garcia-Campello M, Pousa A, Powell A, Baker-Austin C, Gonzalez-Escalona N (2018) Epidemic dynamics of Vibrio parahaemolyticus illness in a hotspot of disease emergence, Galicia, Spain. Emerg Infect Dis 24:852–859
Baker-Austin C, Trinanes JA, Salmenlinna S, Lofdahl M, Siitonen A, Taylor NGH, Martinez-Urtaza J (2016) Heat wave-associated vibriosis, Sweden and Finland, 2014. Emerg Infect Dis 22:1216–1220
Elmahdi S, DaSilva LV, Parveen S (2016) Antibiotic resistance of Vibrio parahaemolyticus and Vibrio vulnificus in various countries: a review. Food Microbiol 57:128–134
Letchumanan V, Chan K-G, Lee L-H (2015) An insight of traditional plasmid curing in Vibrio species. Front Microbiol 6:735
He Y, Jin L, Sun F, Hu Q, Chen L (2016) Antibiotic and heavy-metal resistance of Vibrio parahaemolyticus isolated from fresh shrimps in Shanghai fish markets, China. Environ Sci Pollut Res 23:15033–15040
Zanetti S, Spanu T, Deriu A, Romano L, Sechi LA, Fadda G (2001) In vitro susceptibility of Vibrio spp. isolated from the environment. Int J Antimicrob Agents 17:407–409
Lee L-H, Raghunath P (2018) Editorial: vibrionaceae diversity multidrug resistance and management. Front Microbiol 9:563
Yang Y, Xie J, Li H, Tan S, Chen Y, Yu H (2017) Prevalence, antibiotic susceptibility and diversity of Vibrio parahaemolyticus isolates in seafood from South China. Front Microbiol 8:2566
Lopatek M, Wieczorek K, Osek J (2018) Antimicrobial resistance, virulence factors, and genetic profiles of Vibrio parahaemolyticus from seafood. Appl Environ Microbiol 84:e00537-e1518
Jiang Y, Chu Y, Xie G, Li F, Wang L, Huang J, Zhai Y, Yao L (2019) Antimicrobial resistance, virulence and genetic relationship of Vibrio parahaemolyticus in seafood from coasts of Bohai Sea and Yellow Sea, China. Int J Food Microbiol 290:116–124
Lei T, Jiang F, He M, Zhang J, Zeng H, Chen M, Pang R, Wu S, Wei L, Wang J, Ding Y, Wu Q (2020) Prevalence, virulence, antimicrobial resistance, and molecular characterization of fluoroquinolone resistance of Vibrio parahaemolyticus from different types of food samples in China. Int J Food Microbiol 317:108461
Freire-Moran L, Aronsson B, Manz C, Gyssens IC, So AD, Monnet DL, Cars O, Grp E-EW (2011) Critical shortage of new antibiotics in development against multidrug-resistant bacteria-Time to react is now. Drug Resist Updates 14:118–124
Obaidat MM, Salman AEB, Roess AA (2017) Virulence and antibiotic resistance of Vibrio parahaemolyticus isolates from seafood from three developing countries and of worldwide environmental, seafood, and clinical isolates from 2000 to 2017. J Food Prot 80:2060–2067
Letchumanan V, Chan K-G, Pusparajah P, Saokaew S, Duangjai A, Goh B-H, Ab Mutalib N-S, Lee L-H (2016) Insights into bacteriophage application in controlling Vibrio species. Front Microbiol 7:1114
Zhang H, Yang Z, Zhou Y, Bao H, Wang R, Li T, Pang M, Sun L, Zhou X (2018) Application of a phage in decontaminating Vibrio parahaemolyticus in oysters. Int J Food Microbiol 275:24–31
Yang MY, Liang YJ, Su RB, Chen HF, Wang J, Zhang JM, Ding Y, Kong L, Zeng HY, Xue L, Wu HM, Wu QP (2019) Genome characterization of the novel lytic Vibrio parahaemolyticus phage vB_VpP_BA6. Arch Virol 164:2627–2630
Ding T, Sun H, Pan Q, Zhao F, Zhang Z, Ren H (2020) Isolation and characterization of Vibrio parahaemolyticus bacteriophage vB_VpaS_PG07. Virus Res 286:198080
Li MZ, Jin YQ, Lin H, Wang JX, Jiang XP (2018) Complete genome of a novel lytic Vibrio parahaemolyticus phage VPp1 and characterization of its endolysin for antibacterial activities. J Food Prot 81:1117–1125
Rodela ML, Sabet S, Peterson A, Dillon JG (2019) Broad environmental tolerance for a salicola host-phage pair isolated from the Cargill Solar Saltworks, Newark, CA, USA. Microorganisms 7:106
Sabet S, Diallo L, Hays L, Jung W, Dillon JG (2009) Characterization of halophiles isolated from solar salterns in Baja California, Mexico. Extremophiles 13:643–656
Yaashikaa PR, Saravanan A, Kumar PS (2016) Isolation and identification of Vibrio cholerae and Vibrio parahaemolyticus from prawn (Penaeus monodon) seafood: preservation strategies. Microb Pathog 99:5–13
Hyman P, Abedon ST (2010) Bacteriophage host range and bacterial resistance. Adv Appl Microbiol 70(70):217–248
Hyman P, Abedon ST (2009) Practical methods for determining phage growth parameters. Methods Mol Biol (Clifton, NJ) 501:175–202
Pujato SA, Guglielmotti DM, Martinez-Garcia M, Quiberoni A, Mojica FJM (2017) Leuconostoc mesenteroides and Leuconostoc pseudomesenteroides bacteriophages: Genomics and cross-species host ranges. Int J Food Microbiol 257:128–137
Alagappan KM, Deivasigamani B, Somasundaram ST, Kumaran S (2010) Occurrence of Vibrio parahaemolyticus and its specific phages from shrimp ponds in East Coast of India. Curr Microbiol 61:235–240
Buchanan RE, Gibbons NE, Cowan ST (1994) Bergey’s manual of determinative bacteriology. Williams & Wilkins
Heinemeyer EA (1984) Comparative studies on nutritional and physiological characteristics of Vibrio parahaemolyticus originating from Germany, Togo and Peru. Zentralblatt fur Bakteriologie, Mikrobiologie, und Hygiene Series A, Medical microbiology, infectious diseases, virology, parasitology 256:443–455
Deng Y, Xu L, Chen H, Liu S, Guo Z, Cheng C, Ma H, Feng J (2020) Prevalence, virulence genes, and antimicrobial resistance of Vibrio species isolated from diseased marine fish in South China. Sci Rep 10:14329
Soto-Rodriguez SA, Lozano-Olvera R, Palacios-Gonzalez DA, Bolan-Mejia C, Rendon-Aguilar KG (2019) Characterization and growth conditions of Vibrio parahaemolyticus strains with different virulence degrees that cause acute hepatopancreatic necrosis disease in Litopenaeus vannamei. J World Aquacult Soc 50:1002–1015
Hacker J, Carniel E (2001) Ecological fitness, genomic islands and bacterial pathogenicity—a Darwinian view of the evolution of microbes. EMBO Rep 2:376–381
Tran Thi Hong T, Yanagawa H, Nguyen Khanh T, Du Minh H, Van Doan C, Ly Thi Lien K, Taniguchi T, Kubo R, Hayashidani H (2020) Prevalence of Vibrio parahaemolyticus causing acute hepatopancreatic necrosis disease of shrimp in shrimp, molluscan shellfish and water samples in the Mekong Delta, Vietnam. Biology (Basel) 9:312
Mohamad N, Amal MNA, Saad MZ, Yasin ISM, Zulkiply NA, Mustafa M, Nasruddin NS (2019) Virulence-associated genes and antibiotic resistance patterns of Vibrio spp. isolated from cultured marine fishes in Malaysia. BMC Vet Res 15:176
Matamp N, Bhat SG (2020) Genome characterization of novel lytic Myoviridae bacteriophage phi VP-1 enhances its applicability against MDR-biofilm-forming Vibrio parahaemolyticus. Arch Virol 165:387–396
Matsuzaki S, Inoue T, Tanaka S, Koga T, Kuroda M, Kimura S, Imai S (2000) Characterization of a novel Vibrio parahaemolyticus phage, KVP241, and its relatives frequently isolated from seawater. Microbiol Immunol 44:953–956
Luhtanen A-M, Eronen-Rasimus E, Kaartokallio H, Rintala J-M, Autio R, Roine E (2014) Isolation and characterization of phage–host systems from the Baltic Sea ice. Extremophiles 18:121–130
Yang M, Liang Y, Huang S, Zhang J, Wang J, Chen H, Ye Y, Gao X, Wu Q, Tan Z (2020) Isolation and characterization of the novel phages vB_VpS_BA3 and vB_VpS_CA8 for lysing Vibrio parahaemolyticus. Front Microbiol 11:259
Yu Y-P, Gong T, Jost G, Liu W-H, Ye D-Z, Luo Z-H (2013) Isolation and characterization of five lytic bacteriophages infecting a Vibrio strain closely related to Vibrio owensii. FEMS Microbiol Lett 348:112–119
Abedon ST, Herschler TD, Stopar D (2001) Bacteriophage latent-period evolution as a response to resource availability. Appl Environ Microbiol 67:4233–4241
Wang IN, Dykhuizen DE, Slobodkin LB (1996) The evolution of phage lysis timing. Evol Ecol 10:545–558
Finke JF, Hunt BPV, Winter C, Carmack EC, Suttle CA (2017) Nutrients and other environmental factors influence virus abundances across oxic and hypoxic marine environments. Viruses-Basel 9:152
Sandaa R-A, Gomez-Consarnau L, Pinhassi J, Riemann L, Malits A, Weinbauer MG, Gasol JM, Thingstad TF (2009) Viral control of bacterial biodiversity—evidence from a nutrient-enriched marine mesocosm experiment. Environ Microbiol 11:2585–2597
Zhang D, You F, He Y, Te SH, Gin KY-H (2020) Isolation and characterization of the first freshwater cyanophage infecting Pseudanabaena. J Virol 94:e00682-e1620
Gong Z, Wang M, Yang Q, Li Z, Xia J, Gao Y, Jiang Y, Meng X, Liu Z, Yang D, Zhang F, Shao H, Wang D (2017) Isolation and complete genome sequence of a novel Pseudoalteromonas phage PH357 from the Yangtze River Estuary. Curr Microbiol 74:832–839
Labrie SJ, Samson JE, Moineau S (2010) Bacteriophage resistance mechanisms. Nat Rev Microbiol 8:317–327
Diaz-Munoz SL, Koskella B (2014) Bacteria-phage interactions in natural environments. In: Sariaslani S, Gadd GM (eds) Adv Appl Microbiol, pp 135–183
Dekel-Bird NP, Avrani S, Sabehi G, Pekarsky I, Marston MF, Kirzner S, Lindell D (2013) Diversity and evolutionary relationships of T7-like podoviruses infecting marine cyanobacteria. Environ Microbiol 15:1476–1491
Gentile GM, Wetzel KS, Dedrick RM, Montgomery MT, Garlena RA, Jacobs-Sera D, Hatfull GF (2019) More evidence of collusion: a new prophage-mediated viral defense system encoded by mycobacteriophage Sbash. MBio 10:e00196-e1119
Tanji Y, Hattori K, Suzuki K, Miyanaga K (2008) Spontaneous deletion of a 209-kilobase-pair fragment from the Escherichia coli genome occurs with acquisition of resistance to an assortment of infectious phages. Appl Environ Microbiol 74:4256–4263
Zuo P, Yu P, Alvarez PJJ (2020) Beta-lactam-induced outer membrane alteration confers E. coli a fortuitous competitive advantage through cross-resistance to bacteriophages. Environ Sci Technol Lett 7:428–433
Hu M, Zhang H, Gu D, Ma Y, Zhou X (2020) Identification of a novel bacterial receptor that binds tail tubular proteins and mediates phage infection of Vibrio parahaemolyticus. Emerg Microbes Infect 9:855–867
Oechslin F (2018) Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses (Basel) 10:351
Acknowledgments
This work was supported by the National Key R&D Program of China (grant number 2018YFD0800104), the Special Fund of China (grant number AWS18J004 and 2019-JCJQ-JJ-163), the Natural Science Foundation of Tianjin, China (grant number 17JCZDJC39100 and 19JCYBJC23800), and the National Natural Science Foundation of China (grant number 51808468). We thank Dan Huang for her help in analyzing the genetic information and function of phage, and Kun He, Yanping Yang, and Chunchun Zhang for their assistance in conducting host growth and phage experiments.
Funding
This research was funded by the National Key R&D Program of China (Grant number 2018YFD0800104), the Special Fund of China (Grant number AWS18J004 and 2019-JCJQ-JJ-163), the Natural Science Foundation of Tianjin, China (Grant number 17JCZDJC39100 and 19JCYBJC23800), and the National Natural Science Foundation of China (Grant number 51808468).
Author information
Authors and Affiliations
Contributions
Conceptualization: CG, XY, C Z, CL, SW, XZ, BX, ZS, JW, LL, PY, ZN, and ZQ. Data curation: CG, XY, ZN, and ZQ. Formal analysis: CG, XY, CZ, SW, ZC, HZ, YY, ZN, and ZQ. Funding acquisition: SW, JW, LL, and ZQ. Investigation: CG, XY, CL, SW, XZ, BX, ZS, JW, ZN, and ZQ. Methodology: CG, XY, CZ, SW, XZ, BX, ZS, JW, PY, ZN, and ZQ. Project administration: JW, ZN, and ZQ. Resources: JW and ZQ. Validation: CG, XY, CZ, SW, ZC, HZ, YY, ZN, and ZQ. Writing—original draft: CG, XY, and ZQ. Writing—review and editing: CG, XY, PY, and ZQ. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflicts or competing interests.
Ethical approval
This research complies with all ethical guidelines, and relevant departments and institutions have approved the research.
Additional information
Handling Editor: Johannes Wittmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gao, C., Yang, X., Zhao, C. et al. Characterization of a novel Vibrio parahaemolyticus host-phage pair and antibacterial effect against the host. Arch Virol 167, 531–544 (2022). https://doi.org/10.1007/s00705-021-05278-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05278-6