Abstract
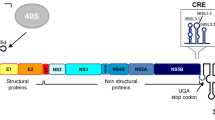
Internal ribosome entry site (IRES)-dependent translation is a mechanism distinct from 5′ cap-dependent translation. IRES elements are located mainly in the 5′ untranslated regions (UTRs) of viral and eukaryotic mRNAs. However, IRESs are also found in the coding regions of some viral and eukaryotic genomes to initiate the translation of some functional truncated isoforms. Here, five putative IRES elements of human rhinovirus 16 (HRV16) were identified in the coding region of the nonstructural proteins P2 and P3 through fusion with green fluorescent protein (GFP) expression vectors and bicistronic vectors with a hairpin structure. These five putative IRESs were located at nucleotide positions 4286-4585, 5002-5126, 6245-6394, 6619-6718, and 6629-6778 in the HRV16 genome. The functionality of the five IRESs was confirmed by their ability to initiate GFP expression in vitro. This suggests that an alternative mechanism might be used to increase the efficiency of replication of HRV16.






Similar content being viewed by others
References
Merrick WC (2004) Cap-dependent and cap-independent translation in eukaryotic systems. Gene 332:1–11. https://doi.org/10.1016/j.gene.2004.02.051
Lindeberg J, Ebendal T (1999) Use of an internal ribosome entry site for bicistronic expression of Cre recombinase or rtTA transactivator. Nucleic Acids Res 27(6):1552–1554. https://doi.org/10.1093/nar/27.6.1552
Ohlmann T, Jackson R (1999) The properties of chimeric picornavirus IRESes show that discrimination between internal translation initiation sites is influenced by the identity of the IRES and not just the context of the AUG codon. RNA (New York, NY) 5(6):764–778. https://doi.org/10.1017/s1355838299982158
Pelletier J, Sonenberg N (1988) Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334(6180):320–325. https://doi.org/10.1038/334320a0
Rojas-Eisenring IA, Cajero-Juarez M, Del Angel RM (1995) Cell proteins bind to a linear polypyrimidine-rich sequence within the 5’-untranslated region of rhinovirus 14 RNA. J Virol 69(11):6819–6824
Yang D, Wilson J, Anderson D, Bohunek L, Cordeiro C, Kandolf R, McManus B (1997) In vitro mutational and inhibitory analysis of the cis-acting translational elements within the 5’ untranslated region of coxsackievirus B3: potential targets for antiviral action of antisense oligomers. Virology 228(1):63–73. https://doi.org/10.1006/viro.1996.8366
Sweeney TR, Abaeva IS, Pestova TV, Hellen CU (2014) The mechanism of translation initiation on Type 1 picornavirus IRESs. EMBO J 33(1):76–92. https://doi.org/10.1002/embj.201386124
Martínez-Salas E, Francisco-Velilla R, Fernandez-Chamorro J, Lozano G, Diaz-Toledano R (2015) Picornavirus IRES elements: RNA structure and host protein interactions. Virus Res 206:62–73. https://doi.org/10.1016/j.virusres.2015.01.012
Lee KM, Chen CJ, Shih SR (2017) Regulation Mechanisms of Viral IRES-Driven Translation. Trends Microbiol 25(7):546–561. https://doi.org/10.1016/j.tim.2017.01.010
Khawaja A, Vopalensky V, Pospisek M (2015) Understanding the potential of hepatitis C virus internal ribosome entry site domains to modulate translation initiation via their structure and function. Wiley Interdiscip Rev RNA 6(2):211–224. https://doi.org/10.1002/wrna.1268
Zhu Z, Wang Y, Yu J, Wan L, Chen J, Xiao M (2010) Classical swine fever virus NS3 is an IRES-binding protein and increases IRES-dependent translation. Virus Res 153(1):106–112. https://doi.org/10.1016/j.virusres.2010.07.013
Xiao M, Wang Y, Zhu Z, Yu J, Wan L, Chen J (2009) Influence of NS5A protein of classical swine fever virus (CSFV) on CSFV internal ribosome entry site-dependent translation. J Gen Virol 90(Pt 12):2923–2928. https://doi.org/10.1099/vir.0.014472-0
Chard LS, Kaku Y, Jones B, Nayak A, Belsham GJ (2006) Functional analyses of RNA structures shared between the internal ribosome entry sites of hepatitis C virus and the picornavirus porcine teschovirus 1 Talfan. J Virol 80(3):1271–1279. https://doi.org/10.1128/JVI.80.3.1271-1279.2006
Chard LS, Bordeleau ME, Pelletier J, Tanaka J, Belsham GJ (2006) Hepatitis C virus-related internal ribosome entry sites are found in multiple genera of the family Picornaviridae. J Gen Virol 87(Pt 4):927–936. https://doi.org/10.1099/vir.0.81546-0
Pfingsten J, Costantino D, Kieft J (2007) Conservation and diversity among the three-dimensional folds of the Dicistroviridae intergenic region IRESes. J Mol Biol 370(5):856–869. https://doi.org/10.1016/j.jmb.2007.04.076
Luo XN, Song QQ, Yu J, Song J, Wang XL, Xia D, Sun P, Han J (2017) Identification of the internal ribosome entry sites (IRES) of prion protein gene. Int J Biochem Cell Biol 93:46–51. https://doi.org/10.1016/j.biocel.2017.10.014
Graber TE, Baird SD, Kao PN, Mathews MB, Holcik M (2010) NF45 functions as an IRES trans-acting factor that is required for translation of cIAP1 during the unfolded protein response. Cell Death Differ 17(4):719–729. https://doi.org/10.1038/cdd.2009.164
Qin X, Sarnow P (2004) Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J Biol Chem 279(14):13721–13728. https://doi.org/10.1074/jbc.M312854200
Song QQ, Luo XN, Chi MM, Song J, Xia D, Han J (2020) Identification of internal ribosomal entry site inside open reading frame of 14–3-3beta gene. Biomed Environ Sci BES 33(4):273–276. https://doi.org/10.3967/bes2020.037
Du X, Gomez CM (2018) Spinocerebellar [corrected] ataxia type 6: molecular mechanisms and calcium channel genetics. Adv Exp Med Biol 1049:147–173. https://doi.org/10.1007/978-3-319-71779-1_7
Dos Santos PN, Canduri F (2018) The emerging picture of CDK11: genetic, functional and medicinal aspects. Curr Med Chem 25(8):880–888. https://doi.org/10.2174/0929867324666170815102036
Khan D, Katoch A, Das A, Sharathchandra A, Lal R, Roy P, Das S, Chattopadhyay S, Das S (2015) Reversible induction of translational isoforms of p53 in glucose deprivation. Cell Death Differ 22(7):1203–1218. https://doi.org/10.1038/cdd.2014.220
Lin JY, Chen TC, Weng KF, Chang SC, Chen LL, Shih SR (2009) Viral and host proteins involved in picornavirus life cycle. J Biomed Sci 16:103. https://doi.org/10.1186/1423-0127-16-103
Fu Y, Cai J, Xi M, He Y, Zhao Y, Zheng Y, Zhang Y, Xi J, He Y (2020) Neuroprotection effect of astragaloside IV from 2-DG-induced endoplasmic reticulum stress. Oxid Med Cell Longev 2020:9782062. https://doi.org/10.1155/2020/9782062
Fang DL, Wan Y, Shen W, Cao J, Sun ZX, Yu HH, Zhang Q, Cheng WH, Chen J, Ning B (2013) Endoplasmic reticulum stress leads to lipid accumulation through upregulation of SREBP-1c in normal hepatic and hepatoma cells. Mol Cell Biochem 381(1–2):127–137. https://doi.org/10.1007/s11010-013-1694-7
Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL (2008) The Vienna RNA websuite. Nucleic Acids Res 36(Web Server issue):W70–W74. https://doi.org/10.1093/nar/gkn188
Ricci EP, Herbreteau CH, Decimo D, Schaupp A, Datta SA, Rein A, Darlix JL, Ohlmann T (2008) In vitro expression of the HIV-2 genomic RNA is controlled by three distinct internal ribosome entry segments that are regulated by the HIV protease and the Gag polyprotein. RNA 14(7):1443–1455. https://doi.org/10.1261/rna.813608
Brasey A, Lopez-Lastra M, Ohlmann T, Beerens N, Berkhout B, Darlix JL, Sonenberg N (2003) The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J Virol 77(7):3939–3949. https://doi.org/10.1128/jvi.77.7.3939-3949.2003
Jackson RJ, Hellen CU, Pestova TV (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11(2):113–127. https://doi.org/10.1038/nrm2838
Sonenberg N, Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136(4):731–745. https://doi.org/10.1016/j.cell.2009.01.042
Jang S, Kräusslich H, Nicklin M, Duke G, Palmenberg A, Wimmer E (1988) A segment of the 5’ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol 62(8):2636–2643. https://doi.org/10.1128/jvi.62.8.2636-2643.1988
Komar AA, Hatzoglou M (2014) Cellular IRES-mediated translation. Cell Cycle 10(2):229–240. https://doi.org/10.4161/cc.10.2.14472
Weingarten-Gabbay S, Elias-Kirma S, Nir R, Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A, Segal E (2016) Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science (New York, NY) 351(6270):4939–4953. https://doi.org/10.1126/science.aad4939
Hanson PJ, Zhang HM, Hemida MG, Ye X, Qiu Y, Yang D (2012) IRES-dependent translational control during virus-induced endoplasmic reticulum stress and apoptosis. Front Microbiol 3:92. https://doi.org/10.3389/fmicb.2012.00092
Jendrach M, Thiel V, Siddell S (1999) Characterization of an internal ribosome entry site within mRNA 5 of murine hepatitis virus. Adv Virol 144(5):921–933. https://doi.org/10.1007/s007050050556
Cornelis S, Bruynooghe Y, Denecker G, Van Huffel S, Tinton S, Beyaert R (2000) Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol Cell 5(4):597–605. https://doi.org/10.1016/s1097-2765(00)80239-7
Wachalska M, Graul M, Praest P, Luteijn RD, Babnis AW, Wiertz E, Bienkowska-Szewczyk K, Lipinska AD (2019) Fluorescent TAP as a platform for virus-induced degradation of the antigenic peptide transporter. Cells 8(12):1590. https://doi.org/10.3390/cells8121590
Chou AC, Aslanian A, Sun H, Hunter T (2019) An internal ribosome entry site in the coding region of tyrosyl-DNA phosphodiesterase 2 drives alternative translation start. J Biol Chem 294(8):2665–2677. https://doi.org/10.1074/jbc.RA118.006269
Thiel V, Siddell SG (1994) Internal ribosome entry in the coding region of murine hepatitis virus mRNA 5. J Gener Virol. https://doi.org/10.1099/0022-1317-75-11-3041
Wang G, Guo X, Silveyra P, Kimball SR, Floros J (2009) Cap-independent translation of human SP-A 5’-UTR variants: a double-loop structure and cis-element contribution. Am J Physiol Lung Cell Mol Physiol 296(4):L635-647. https://doi.org/10.1152/ajplung.90508.2008
Stoneley M, Willis AE (2004) Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene 23(18):3200–3207. https://doi.org/10.1038/sj.onc.1207551
Buck CB, Shen X, Egan MA, Pierson TC, Walker CM, Siliciano RF (2001) The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J Virol 75(1):181–191. https://doi.org/10.1128/JVI.75.1.181-191.2001
Herbreteau CH, Weill L, Decimo D, Prevot D, Darlix JL, Sargueil B, Ohlmann T (2005) HIV-2 genomic RNA contains a novel type of IRES located downstream of its initiation codon. Nat Struct Mol Biol 12(11):1001–1007. https://doi.org/10.1038/nsmb1011
Funding
This work was supported by China Mega-Project for Infectious Disease [2018ZX10102001, 2018ZX10711001, 2018ZX10734401 and 2018ZX10734404], SKLID Development Grant (2011SKLID104) and Project of National Pathogen Resource Center (NPRC-32).
Author information
Authors and Affiliations
Contributions
Jun Han designed and conceived the experiments and analyzed the data; Bingtian Shi performed the experiments, analyzed data, and wrote the original draft; Qinqin Song, Xiaonuan Luo, Juan Song, Dong Xia, Zhiqiang Xia, Mi Liu, Wenjun Wang, Ruifang Wang, and Haijun Du supervised data; Jun Han revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Handling Editor: Ana Cristina Bratanich.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shi, B., Song, Q., Luo, X. et al. Identification of cryptic putative IRESs within the ORF encoding the nonstructural proteins of the human rhinovirus 16 genome. Arch Virol 166, 3373–3386 (2021). https://doi.org/10.1007/s00705-021-05209-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05209-5