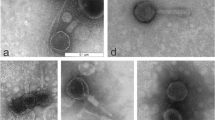
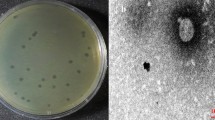
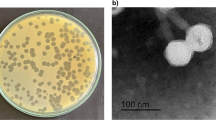
Abstract
Antimicrobial resistance is a serious threat to public health around the globe. According to the World Health Organization, there will be a return to the pre-penicillin era by 2050 if no new antimicrobials are discovered. It is therefore necessary to find new antimicrobials and alternatives. Pseudomonas aeruginosa exhibits resistance against many antibiotics and causes a variety of infections in immunocompromised individuals and especially in those with burn wounds and lung infections. Bacteriophage RLP against P. aeruginosa strain PA-1 was isolated from the Ravi River near Lahore. It showed marked stability at different pH values and temperatures, with the maximum storage stability at 4 °C. It demonstrated the ability to inhibit bacterial growth for up to 20 h, replicated in 25 min, and produced 154 virions per infected cell. RLP showed a broad host range, infecting 50% (19/38) of the multiple-drug-resistant (MDR) P. aeruginosa strains that were tested. The 43-kbp-long genome of RLP is a double-stranded DNA molecule that encodes 56 proteins in total: 34 with known functions, and 22 with no homolog in the gene databases. A cascade system of lytic machinery is also present in the form of four genes (R/z, R/z1, holin and endolysin). Therapeutic studies of RLP in bacteremic mice infected with P. aeruginosa strain PA-1 demonstrated a 92% survival rate in the treated group compared with 7.4% in the untreated group, and this result was statistically significant. Based on its physiological and genetic properties, ability to cause a reduction in bacterial growth in vitro and its in vivo therapeutic efficacy, RLP could be a good candidate for use in phage therapy.






Similar content being viewed by others
References
Church D, Elsayed S, Reid O, Winston B, Lindsay R (2006) Burn wound infections. Clin Microbiol Rev 19:403–434
Pier G, Ramphal R (2005) Pseudomonas aeruginosa. Princip Pract Infect Dis 6:2587–2615
McVay CS, Velásquez M, Fralick JA (2007) Phage therapy of Pseudomonas aeruginosa infection in a mouse burn wound model. Antimicrob Agents Chemother 51:1934–1938
Merril CR, Scholl D, Adhya SL (2003) The prospect for bacteriophage therapy in Western medicine. Nat Rev Drug Discov 2:489
Brüssow H (2005) Phage therapy: the Escherichia coli experience. Microbiology 151:2133–2140
Barrow PA, Soothill JS (1997) Bacteriophage therapy and prophylaxis: rediscovery and renewed assessment of potential. Trends Microbiol 5:268–271
Smith HW, Huggins M (1983) Effectiveness of phages in treating experimental Escherichia coli diarrhoea in calves, piglets and lambs. Microbiology 129:2659–2675
Pirisi A (2000) Phage therapy—advantages over antibiotics? Lancet 356:1418
Stone R (2002) Stalin’s forgotten cure. Science 298:728–731
Gorski A, Dabrowska K, Switala-Jeleń K, Nowaczyk M, Weber-Dabrowska B, Boratynski J, Wietrzyk J, Opolski A (2003) New insights into the possible role of bacteriophages in host defense and disease. Med Immunol 2:2
Kochetkova VA, Mamontov AS, Moskovtseva RL, Erastova EI, Trofimov EI, Popov MI, Dzhubalieva SK (1989) Phagotherapy of postoperative suppurative-inflammatory complications in patients with neoplasms. Sovetskaia meditsina (6), 23–26
Sulakvelidze A, Alavidze Z, Morris JG (2001) Bacteriophage therapy. Antimicrob Agents Chemother 45:649–659
Yin S, Huang G, Zhang Y, Jiang B, Yang Z, Dong Z, You B, Yuan Z, Hu F, Zhao Y (2017) Phage Abp1 rescues human cells and mice from infection by pan-drug resistant Acinetobacter baumannii. Cell Physiol Biochem 44:2337–2345
Khawaja KA, Rauf M, Abbas Z (2016) A virulent phage JHP against Pseudomonas aeruginosa showed infectivity against multiple genera. J Basic Microbiol 56:1090–1097
Bibi Z, Abbas Z, Rehman Su (2016) The phage P. E1 isolated from hospital sewage reduces the growth of Escherichia coli. Biocontrol Sci Tech 26:181–188
Kutter E (2009) Phage host range and efficiency of plating. Bacteriophages. Springer, Berlin, pp 141–149
Alvi IA, Asif M, Tabassum R, Abbas Z, ur Rehman S (2018) Storage of bacteriophages at 4 C leads to no loss in their titer after one year. Pakis J Zool 50:1999–2398
Ellis EL, Delbruck M (1939) The growth of bacteriophage. J Gen Physiol 22:365–384
Asif M, Alvi IA, Tabassum R, Rehman SU (2020) TAC1, an unclassified bacteriophage of the family Myoviridae infecting Acinetobacter baumannii with a large burst size and a short latent period. Arch Virol 165(2):419–424
Sambrook J, Russell DW (2006) Purification of nucleic acids by extraction with phenol: chloroform. Cold Spring Harbor Protocols 2006:pdb. prot4455
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genom 9:75
Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS (2016) PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–W21
Lavigne R, Sun W, Volckaert G (2004) PHIRE, a deterministic approach to reveal regulatory elements in bacteriophage genomes. Bioinformatics 20:629–635
Tiwari BR, Kim S, Rahman M, Kim J (2011) Antibacterial efficacy of lytic Pseudomonas bacteriophage in normal and neutropenic mice models. J Microbiol 49:994–999
Skorupski K, Pierce JC, Sauer B, Sternberg N (1992) Bacteriophage P1 genes involved in the recognition and cleavage of the phage packaging site (pac). J Mol Biol 223:977–989
Pallavali RR, Degati VL, Lomada D, Reddy MC, Durbaka VRP (2017) Isolation and in vitro evaluation of bacteriophages against MDR-bacterial isolates from septic wound infections. PLoS One 12:e0179245
Haq IU, Chaudhry WN, Andleeb S, Qadri I (2012) Isolation and partial characterization of a virulent bacteriophage IHQ1 specific for Aeromonas punctata from stream water. Microb Ecol 63:954–963
Ceyssens P-J, Lavigne R, Mattheus W, Chibeu A, Hertveldt K, Mast J, Robben J, Volckaert G (2006) Genomic analysis of Pseudomonas aeruginosa phages LKD16 and LKA1: establishment of the φKMV subgroup within the T7 supergroup. J Bacteriol 188:6924–6931
Jamal M, Chaudhry WN, Hussain T, Das CR, Andleeb S (2015) Characterization of new Myoviridae bacteriophage WZ1 against multi-drug resistant (MDR) Shigella dysenteriae. J Basic Microbiol 55:420–431
Jończyk E, Kłak M, Międzybrodzki R, Górski A (2011) The influence of external factors on bacteriophages. Folia Microbiol 56:191–200
Ackermann HW, Tremblay D, Moineau S (2004) Long-term bacteriophage preservation. WFCC Newslett 38:35–40
Garbe J, Wesche A, Bunk B, Kazmierczak M, Selezska K, Rohde C, Sikorski J, Rohde M, Jahn D, Schobert M (2010) Characterization of JG024, a Pseudomonas aeruginosa PB1-like broad host range phage under simulated infection conditions. BMC Microbiol 10:301
Ahiwale S, Prakash D, Gajbhiye M, Jagdale S, Patil N, Kapadnis B (2012) BVPaP-3, a T7-like lytic phage of Pseudomonas aeruginosa: its isolation and characterisation. Curr Microbiol 64:305–311
Lavigne R, Burkal’tseva MV, Robben J, Sykilinda NN, Kurochkina LP, Grymonprez B, Jonckx B, Krylov VN, Mesyanzhinov VV, Volckaert G (2003) The genome of bacteriophage φKMV, a T7-like virus infecting Pseudomonas aeruginosa. Virology 312:49–59
Yamaguchi K, Miyata R, Shigehisa R, Uchiyama J, Takemura-Uchiyama I, Kato S-I, Ujihara T, Sakaguchi Y, Daibata M, Matsuzaki S (2014) Genome analysis of Pseudomonas aeruginosa bacteriophage KPP23, belonging to the family Siphoviridae. Genome Announc 2:e00233–e00234
Krylov V, Pleteneva E, Bourkaltseva M, Shaburova O, Volckaert G, Sykilinda N, Kurochkina L, Mesyanzhinov V (2003) Myoviridae bacteriophages of Pseudomonas aeruginosa: a long and complex evolutionary pathway. Res Microbiol 154:269–275
Alves DR, Perez-Esteban P, Kot W, Bean J, Arnot T, Hansen LH, Enright MC, Jenkins ATA (2016) A novel bacteriophage cocktail reduces and disperses Pseudomonas aeruginosa biofilms under static and flow conditions. Microb Biotechnol 9:61–74
Karumidze N, Thomas JA, Kvatadze N, Goderdzishvili M, Hakala KW, Weintraub ST, Alavidze Z, Hardies SC (2012) Characterization of lytic Pseudomonas aeruginosa bacteriophages via biological properties and genomic sequences. Appl Microbiol Biotechnol 94:1609–1617
Essoh C, Latino L, Midoux C, Blouin Y, Loukou G, Nguetta S-PA, Lathro S, Cablanmian A, Kouassi AK, Vergnaud G (2015) Investigation of a large collection of Pseudomonas aeruginosa bacteriophages collected from a single environmental source in Abidjan, Côte d’Ivoire. PloS One 10:e0130548
Streisinger G, Emrich J, Stahl MM (1967) Chromosome structure in phage t4, iii. Terminal redundancy and length determination. Proc Natl Acad Sci 57:292–295
Ceyssens P-J, Hertveldt K, Ackermann H-W, Noben J-P, Demeke M, Volckaert G, Lavigne R (2008) The intron-containing genome of the lytic Pseudomonas phage LUZ24 resembles the temperate phage PaP3. Virology 377:233–238
Hanych B, Kędzierska S, Walderich B, Uznański B, Taylor A (1993) Expression of the Rz gene and the overlapping Rz1 reading frame present at the right end of the bacteriophage lambda genome. Gene 129:1–8
Kȩdzierska S, Wawrzynow A, Taylor A (1996) The Rz1 gene product of bacteriophage lambda is a lipoprotein localized in the outer membrane of Escherichia coli. Gene 168:1–8
Young R, Wang N, Roof WD (2000) Phages will out: strategies of host cell lysis. Trends Microbiol 8:120–128
Berry J, Summer EJ, Struck DK, Young R (2008) The final step in the phage infection cycle: the Rz and Rz1 lysis proteins link the inner and outer membranes. Mol Microbiol 70:341–351
Grymonprez B, Jonckx B, Krylov V, Mesyanzhinov V, Volckaert G (2003) The genome of bacteriophage phiKMV, a T7-like virus infecting Pseudomonas aeruginosa. Virology 312:4959
Cao Z, Zhang J, Niu YD, Cui N, Ma Y, Cao F, Jin L, Li Z, Xu Y (2015) Isolation and characterization of a “phiKMV-like” bacteriophage and its therapeutic effect on mink hemorrhagic pneumonia. PLoS One 10:e0116571
Payne RJ, Jansen VA (2003) Pharmacokinetic principles of bacteriophage therapy. Clin Pharmacokinet 42:315–325
Payne RJ, Jansen VA (2000) Phage therapy: the peculiar kinetics of self-replicating pharmaceuticals. Clin Pharmacol Ther 68:225–230
Uchiyama J, Maeda Y, Takemura I, Chess-Williams R, Wakiguchi H, Matsuzaki S (2009) Blood kinetics of four intraperitoneally administered therapeutic candidate bacteriophages in healthy and neutropenic mice. Microbiol Immunol 53:301–304
Schneider G, Szentes N, Horváth M, Dorn Á, Cox A, Nagy G, Dofkay Z, Maróti G, Rákhely G, Kovács T (2018) Kinetics of targeted phage rescue in a mouse model of systemic Escherichia coli K1. BioMed Res Int 2018:7569645
Henry M, Lavigne R, Debarbieux L (2013) Predicting in vivo efficacy of therapeutic bacteriophages used to treat pulmonary infections. Antimicrob Agents Chemother 57:5961–5968
Author information
Authors and Affiliations
Contributions
The research work was conducted as a part of the PhD research of Iqbal Ahmad Alvi. Shafiq Ur Rehman and Iqbal Ahmad Alvi designed the study; Iqbal Ahmad Alvi and Muhammad Asif performed the in vitro studies; Iqbal Ahmad Alvi, Rehan Aslam and Zaigham Abbas performed animal model studies; and Iqbal Ahmad Alvi, Muhammad Asif, Rabia Tabassum and Shafiq Ur Rehman perfomed the pharmacokinetics studies, compiled the data and prepared the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Handling Editor: T. K. Frey.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The 16S rRNA sequence of the bacterial strain (Pseudomonas aeruginosa strain PA-1) was submitted to NCBI GenBank with accession number MG763232. The whole genome sequence of bacteriophage RLP was submitted to NCBI GenBank with accession number MH979674.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Alvi, I.A., Asif, M., Tabassum, R. et al. RLP, a bacteriophage of the family Podoviridae, rescues mice from bacteremia caused by multi-drug-resistant Pseudomonas aeruginosa. Arch Virol 165, 1289–1297 (2020). https://doi.org/10.1007/s00705-020-04601-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04601-x