Abstract
Pigeon circovirus (PiCV) is taxonomically classified as a member of the Circovirus genus, family Circoviridae. The virus contains a single stranded DNA genome of approximately 2 kb, with minor length variations among different isolates. The occurrence of PiCV infections in pigeons (Columba livia) has been documented worldwide over the past 20 years; however, in Brazil there were still no reports on PiCV detection. This study identifies seven PiCV genomes recovered from domestic pigeons of South Brazil through high-throughput sequencing and shows a high frequency of PiCV infection, through quantitative real-time PCR. Phylogenetic classification was performed by maximum likelihood analysis of the full genomes, ORF V1 (Rep) and ORF C1 (Cap). The results show that either full genome or Cap based analysis allowed PiCV classification into five major clades (groups A to E), where Brazilian sequences were classified as A, C or D. Recombination analyses were carried out with Simplot and RDP4 and the results show that both Rep and Cap ORFs contain several recombination hotspots, pointing to an important role for such events in PiCV evolution.



Similar content being viewed by others
Change history
24 August 2018
Unfortunately, the word “evolution” was found missing in title of the original article which is corrected here by this erratum. The original article has been corrected.
References
Mankertz A, Hattermann K, Ehlers B, Soike D (2000) Cloning and sequencing of columbid circovirus (CoCV), a new circovirus from pigeons. Arch Virol 145:2469–2479. https://doi.org/10.1007/s007050070002
Duchatel JP, Todd D, Curry A et al (2005) New data on the transmission of pigeon circovirus. Vet Rec 157:413–415. https://doi.org/10.1136/vr.157.14.413
Rosario K, Breitbart M, Harrach B et al (2017) Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch Virol 162:1447–1463. https://doi.org/10.1007/s00705-017-3247-y
Stenzel T, Farkas K, Varsani A (2015) Genome sequence of a diverse goose circovirus recovered from greylag goose. Genome Announc. https://doi.org/10.1128/genomeA.00767-15
Cságola A, Lőrincz M, Tombácz K et al (2012) Genetic diversity of pigeon circovirus in Hungary. Virus Genes 44:75–79. https://doi.org/10.1007/s11262-011-0669-6
Yamamoto E, Ito H, Kitamoto E et al (2015) Complete genome sequence of pigeon circovirus detected in racing pigeons in western Japan. Virus Genes 51:140–143. https://doi.org/10.1007/s11262-015-1211-z
Delwart E, Li L (2012) Rapidly expanding genetic diversity and host range of the Circoviridae viral family and other Rep encoding small circular ssDNA genomes. Virus Res 164:114–121. https://doi.org/10.1016/j.virusres.2011.11.021
Todd D, Weston JH, Soike D, Smyth JA (2001) Genome sequence determinations and analyses of novel circoviruses from goose and pigeon. Virology 286:354–362. https://doi.org/10.1006/viro.2001.0985
Woods LW, Latimer KS, Niagro FD et al (1994) A retrospective study of circovirus infection in pigeons: nine cases (1986–1993). J Vet Diagn Invest 6:156–164. https://doi.org/10.1177/104063879400600205
Julian L, Piasecki T, Chrzastek K et al (2013) Extensive recombination detected among beak and feather disease virus isolates from breeding facilities in Poland. J Gen Virol 94:1086–1095. https://doi.org/10.1099/vir.0.050179-0
Ng TFF, Willner DL, Lim YW et al (2011) Broad surveys of DNA viral diversity obtained through viral metagenomics of mosquitoes. PLoS ONE. https://doi.org/10.1371/journal.pone.0020579
Dean FB (2001) Rapid amplification of plasmid and phage DNA using Phi29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res 11:1095–1099. https://doi.org/10.1101/gr.180501
Lasken RS (2009) Genomic DNA amplification by the multiple displacement amplification (MDA) method: figure 1. Biochem Soc Trans 37:450–453. https://doi.org/10.1042/BST0370450
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Schmidt HA, Petzold E, Vingron M, von Haeseler A (2003) Molecular phylogenetics: parallelized parameter estimation and quartet puzzling. J Parallel Distrib Comput 63:719–727. https://doi.org/10.1016/S0743-7315(03)00129-1
Guindon S, Dufayard J-F, Lefort V et al (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. https://doi.org/10.1093/sysbio/syq010
Lefort V, Longueville J-E, Gascuel O (2017) SMS: smart model selection in PhyML. Mol Biol Evol 34:2422–2424. https://doi.org/10.1093/molbev/msx149
Anisimova M, Gascuel O (2006) Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol 55:539–552. https://doi.org/10.1080/10635150600755453
Rambaut A (2009) FigTree v1.4: tree figure drawing tool [Internet]
Yu G, Smith DK, Zhu H et al (2017) GGTREE: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36. https://doi.org/10.1111/2041-210X.12628
Muhire B, Martin DP, Brown JK et al (2013) A genome-wide pairwise-identity-based proposal for the classification of viruses in the genus Mastrevirus (family Geminiviridae). Arch Virol 158:1411–1424. https://doi.org/10.1007/s00705-012-1601-7
Lole KS, Bollinger RC, Paranjape RS et al (1999) Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160
Sambrook J, Russel DW (2001) Molecular cloning—a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York
Stenzel T, Piasecki T, Chrzstek K et al (2014) Pigeon circoviruses display patterns of recombination, genomic secondary structure and selection similar to those of beak and feather disease viruses. J Gen Virol 95:1338–1351. https://doi.org/10.1099/vir.0.063917-0
Bencke GA (2007) POMBOS-DOMÉSTICOS Sugestões para o controle em escolas públicas estaduais. http://www.FzbRsGovBr/Upload/20150514114242Pombos_DomesticosPdf. 23
Fontoura PM, Dyer E, Blackburn TM, Orsi ML (2013) Non-native bird species in Brazil Espécies. Neotrop Biol Conserv 8:165–175. https://doi.org/10.4013/nbc.2013.83.07
Duchatel JP, Todd D, Smyth JA et al (2006) Observations on detection, excretion and transmission of pigeon circovirus in adult, young and embryonic pigeons. Avian Pathol 35:30–34. https://doi.org/10.1080/03079450500465692
Stenzel TA, Pestka D, Tykałowski B et al (2012) Epidemiological investigation of selected pigeon viral infections in Poland. Vet Rec 171:562. https://doi.org/10.1136/vr.100932
Wang K-C, Zhuang Q, Qiu Y et al (2017) Genome sequence characterization of pigeon circoviruses in China. Virus Res 233:1–7. https://doi.org/10.1016/j.virusres.2017.03.007
Lefeuvre P, Lett J-M, Varsani A, Martin DP (2009) Widely conserved recombination patterns among single-stranded DNA viruses. J Virol 83:2697–2707. https://doi.org/10.1128/JVI.02152-08
Varsani A, de Villiers GK, Regnard GL et al (2010) A unique isolate of beak and feather disease virus isolated from budgerigars (Melopsittacus undulatus) in South Africa. Arch Virol 155:435–439. https://doi.org/10.1007/s00705-010-0589-0
Lemey P, Posada D (2009) Introduction to recombination detection. In: Lemey P, Salemi M, Vandamme A-M (eds) The phylogenetic handbook, 2nd edn. Cambridge University Press, Cambridge, pp 493–517
Acknowledgements
The authors are grateful to Mr. Luis Gustavo Trainini, who kindly gave access to the samples. This work was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Financiadora de Estudos e Projetos (FINEP, contract 01.10.0783.00). P.M.R. has a CNPq 1A research fellow. This work was carried out as part of the doctoral studies of M.R.L (Programa de Pós-graduação em Ciências Veterinárias, UFRGS).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Handling Editor: Sheela Ramamoorthy.
The original version of this article was revised: The word “evolution” was found missing in title of the original article and is corrected here.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig.S1.
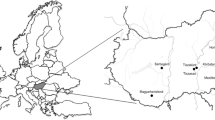
Place of origin of the free-living pigeons used in this study. The points marked on the map are the cities where the pigeons were captured. (DOCX 201 kb)
Fig.S2.
Likelihood mapping of five datasets comprising 82 PiCV sequences. A) Complete genome nucleotide sequences, B) Cap nucleotide sequences, C) Cap amino acid sequences, D) Rep nucleotide sequences, and E) Rep amino acid sequences. (DOCX 395 kb)
Fig. S3.
Bootscanning plots of six PiCV recombinant sequences. Bootscan analyses were performed on a sliding window of 200 nt by increments of 20 nt. A) Genotype A sequence showing no recombination profile; B) Genotype A (Table S1) sequence showing a recombination pattern around positions 1619-1826; C) Genotype A, break point positions around 1480-1826; D) Genotype C sequence presenting no recombination profile; E) and F) Recombination profiles of the sequences from Brazil in this study (genotype C*). (DOCX 818 kb)
Fig.S4.
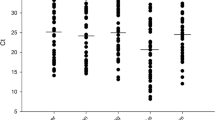
Box plot of PiCV viral genome loads and the city of origin of sampled pigeons. (DOCX 136 kb)
Rights and permissions
About this article
Cite this article
Loiko, M.R., Junqueira, D.M., Varela, A.P.M. et al. Columbid circoviruses detected in free ranging pigeons from Southern Brazil: insights on PiCV evolution. Arch Virol 163, 3083–3090 (2018). https://doi.org/10.1007/s00705-018-3990-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-018-3990-8