Abstract
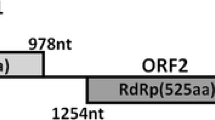
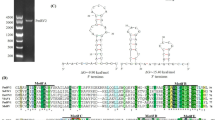
In this study, we describe a novel mycovirus isolated from Ustilaginoidea virens strain GZ-2, which was designated “Ustilaginoidea virens nonsegmented virus 2” (UvNV-2). The genome of UvNV-2 contains two overlapping open reading frames (ORFs). ORF1 encodes an unknown protein, and ORF2 encodes a putative RNA-dependent RNA polymerase (RdRp), which is most closely related to that of Purpureocillium lilacinum nonsegmented virus 1 (PINV-1) and is likely to be expressed by a + 1 ribosomal frameshift within the sequence CCC_UUU_UAG. A phylogenetic analysis of the RdRp of UvNV-2 showed that UvNV-2 is an unclassified mycovirus.


Similar content being viewed by others
References
Mertens P (2004) The dsRNA viruses. Virus Res 101:3–13
Pearson MN, Beever RE, Boine B, Arthur K (2009) Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol Plant Pathol 10:115–128
Ghabrial SA, Suzuki N (2008) Fungal viruses. Encycl Virol 2:284–291
Ghabrial SA, Casto´n JR, Jiang D, Nibert ML, Suzuki N (2015) 50-plus years of fungal viruses. Virology 479:356–368
Zhou Q, Zhong J, Hu Y, Gao BD (2016) A novel nonsegmented double-stranded RNA mycovirus identified in the phytopathogenic fungus Nigrospora oryzae shows similarity to partitivirus-like viruses. Arch Virol 161:229–232
Herrero N (2016) A novel monopartite dsRNA virus isolated from the entomopathogenic and nematophagous fungus Purpureocillium lilacinum. Arch Virol 161:3375–3384
Zhang T, Jiang Y, Dong W (2014) A novel monopartite dsRNA virus isolated from the phytopathogenic fungus Ustilaginoidea virens and ancestrally related to a mitochondria-associated dsRNA in the green alga Bryopsis. Virology 462:227–235
Sun X, Kang S, Zhang Y, Tan X, Yu Y, He H, Zhang X, Liu Y, Wang S, Sun W (2013) Genetic diversity and population structure of rice pathogen Ustilaginoidea virens in China. PLoS One 8:e76879
Tanaka E, Ashizawa T, Sonoda R, Tanaka C (2008) Villosiclava virens gen. nov., comb. nov., the teleomorph of Ustilaginoidea virens, the causal agent of rice false smut. Mycotaxon 106:491–501
Zhang T, Jiang Y, Huang J, Dong W (2013) Complete genome sequence of a putative novel victorivirus from Ustilaginoidea virens. Arch Virol 158:1403–1406
Zhang T, Jiang Y, Huang J, Dong W (2013) Genomic organization of a novel partitivirus from the phytopathogenic fungus Ustilaginoidea virens. Arch Virol 158:2415–2419
Zhong J, Zhu JZ, Lei XH, Chen D, Zhu HJ, Gao BD (2014) Complete genome sequence and organization of a novel virus from the rice false smut fungus Ustilaginoidea virens. Virus Genes 48:329–333
Jiang Y, Zhang T, Luo C, Jiang D, Li G, Li Q, Hsiang T, Huang J (2015) Prevalence and diversity of mycoviruses infecting the plant pathogen Ustilaginoidea virens. Virus Res 195:47–56
Zhong J, ChengYC Gao BD, Zhou Q, Zhu HJ (2017) Mycoviruses in the plant pathogen Ustilaginoidea virens are not correlated with the genetic backgrounds of its hosts. Int J Mol Sci 18:963
Dodds JA, Morris TJ (1983) Double-stranded RNA from plants infected with closteroviruses. Phytopathology 73:419–423
Froussard P (1992) A random-PCR method (rPCR) to construct whole cDNA library from low amounts of RNA. Nucleic Acids Res 20:2900
Lambden PR, Cooke SJ, Caul EO, Clarke IN (1992) Cloning of noncultivatable human rotavirus by single primer amplification. J Virol 66:1817–1822
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG, Valentin F (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Firth A, Jagger B, Wise H, Nelson C, Parsawar K, Wills N, Napthine S, Taubenberger J, Digard P, Atkins J (2012) Ribosomal frameshifting used in influenza A virus expression occurs within the sequence UCC_UUU_CGU and is in the +1 direction. Open Biol. https://doi.org/10.1098/rsob.120109
Agranovsky A, Koonin EV, Boyko VP, Maiss E, Frötschl R, Lunina NA, Atabekov JG (1994) Beet yellows closterovirus: complete genome structure and identification of a leader papainlike thiol protease. Virology 198:311–324
Klobutcher LA, Farabaugh PJ (2002) Shifty ciliates. Frequent programmed translational frameshifting in euplotids. Cell 111:763–766
Weiss RB, Dunn DM, Atkins JF, Gesteland RF (1987) Slippery runs, shifty stops, backward steps, and forward hops: − 2, − 1, + 1, + 2, + 5, and + 6 ribosomal frameshifting. Cold Spring Harb Symp Quant Biol 52:687–693
Koga R, Fukuhara T, Nitta T (1998) Molecular characterization of a single mitochondria-associated double-stranded RNA in the green alga Bryopsis. Plant Mol Biol 36:717–724
Shi M, Neville P, Nicholson J, Eden JS, Imrie A, Holmes EC (2017) High resolution meta-transcriptomics reveals the ecological dynamics of mosquito-associated RNA viruses in Western Australia. J Virol. https://doi.org/10.1128/JVI.00680-17
Funding
This study was funded by grants from the National Natural Science Foundation of China (31760015); the Science and Technology Foundation of Guizhou Province ([2017]1153); the Joint Foundation of Collaboration Project between Scientific and Technological Bureau of Guizhou Province and Universities of Guizhou Province (LH[2015]7322); the Bureau of Science and Technology of Guiyang ([2017]5-24); the Doctoral Cultivating Fund of Guizhou Medical University (J[2014]025); and the Engineering Research Center of Medical Biotechnology, Guizhou Medical University (2016002).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with animals or human participants performed by any of the authors.
Additional information
Handling Editor: Robert H. A. Coutts.
Rights and permissions
About this article
Cite this article
Zhang, T., Zeng, X., Zeng, Z. et al. A novel monopartite dsRNA virus isolated from the phytopathogenic fungus Ustilaginoidea virens strain GZ-2. Arch Virol 163, 3427–3431 (2018). https://doi.org/10.1007/s00705-018-3976-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-018-3976-6