Abstract
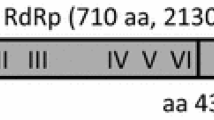
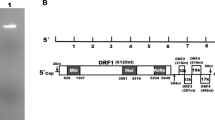
Double-stranded RNAs from an isolate of Neofusicoccum luteum collected from grapevines were analyzed by high-throughput sequencing. Contig annotations revealed the presence of a potential novel virus belonging to the newly proposed family Fusariviridae. Completion of the viral genome sequence was performed. The genome is 6,244 nucleotide long, excluding the poly(A) tail and contains two putative open reading frames (ORFs). The first one encodes a large polypeptide of 1,552 amino acids (aa) with conserved RNA-dependent RNA polymerase and helicase domains typical of viral replicases. The second ORF encodes a putative 475-aa-long polypeptide showing weak homology to the corresponding ORF of Macrophomina phaseolina single-stranded RNA virus 1, for which no function is known so far. Phylogenetic analyses indicated that this virus should be considered a novel mycovirus belonging to the proposed family Fusariviridae, for which the name “Neofusicoccum luteum fusarivirus 1” (NlFV1) is proposed.

Similar content being viewed by others
References
Wunderlich N, Ash G, Steel C, Rahman H, Savocchia S (2010) Botryosphaeriaceae associated with bunch rot of grapes in South Eastern Australia. Phytopathol Mediterr 49:106
Yacoub A, Gerbore J, Magnin N, Chambon P, Dufour MC, Corio-Costet MF, Guyoneaud P, Rey P (2016) Ability of Pythium oligandrum strains to protect Vitis vinifera against Phaeomoniella chlamydospora, a pathogen involved in Esca, by inducing plant resistance. Biol Control 92:7–16
Ghabrial SA, Castón JR, Jiang D, Nibert ML, Suzuki N (2015) 50-plus years of fungal viruses. Virology 479–480:356–368
Wang LP, Jiang JJ, Wang YF, Hong N, Zhang F, Xu WX, Wang GP (2014) Hypovirulence of the phytopathogenic fungus Botryosphaeria dothidea: association with a coinfecting Chrysovirus and a Partitivirus. J Virol 88:7517–7527
Zhai L, Xiang J, Zhang M, Fu M, Yang Z, Hong N, Wang G (2016) Characterization of a novel double-stranded RNA mycovirus conferring hypovirulence from the phytopathogenic fungus Botryosphaeria dothidea. Virology 493:75–85
Phillips AJL (2002) Botryosphaeria species associated with diseases of grapevines in Portugal. Phytopathol Mediterr 41:3–18
Marais A, Nivault A, Faure C, Theil S, Comont G, Candresse T, Corio-Costet MF (2017) Determination of the complete genomic sequence of Neofusicoccum luteum mitovirus 1 (NLMV1), a novel mitovirus associated with a phytopathogenic Botryosphaeriaceae. Arch Virol 162:2477–2480
Zhang R, Liu S, Chiba S, Kondo H, Kanematsu S, Suzuki N (2014) A novel single-stranded RNA virus isolated from a phytopathogenic filamentous fungus, Rosellinia necatrix, with similarity to hypo-like viruses. Front Microbiol 5:e360
Zhong J, Zhao SQ, Li GF, Pang XD, Deng XJ, Zhu HJ, Gao BD, Zhang Q (2016) A novel fusarivirus isolated from the phytopathogenic fungus Nigrospora oryzae. Virus Genes 52:891–895
Bellée A, Comont G, Nivault A, Abou-Mansour E, Coppin C, Dufour MC, Corio-Costet MF (2017) Life traits of four Botryosphaeriacea species and molecular responses of different genotypes and cultivar of Vitis vinifera. Plant Pathol 66:763–776
Marzano SL, Nelson BD, Ajayi-Oyetunde O, Bradley CA, Hughes TJ, Hartman GL, Eastburn DM, Domier LL (2016) Identification of diverse mycoviruses through metatranscriptomics: characterization of the viromes of five major fungal plant pathogens. J Virol 90:6846–6863
Liu R, Cheng J, Fu Y, Jiang D, Xie J (2015) Molecular characterization of a novel positive-sense, single-stranded RNA mycovirus infecting the plant pathogenic fungus Sclerotinia sclerotiorum. Viruses 7:2470–2484
Zhong J, Shang HH, Zhu CX, Zhu JZ, Zhu HJ, Hua Y, Gao BD (2016) Characterization of a novel single-stranded RNA virus, closely related to fusariviruses, infecting the plant pathogenic fungus Alternaria brassicicola. Virus Res 217:1–7
Chu YM, Jeon JJ, Yea SJ, Kim YH, Yun SH, Lee YW, Kim KH (2002) Double-stranded RNA mycovirus from Fusarium graminearum. Appl Environ Microbiol 68:2529–2534
Nerva L, Ciuffo M, Vallino M, Margaria P, Varese GC, Gnavi G, Turina M (2016) Multiple approaches for the detection and characterization of viral and plasmid symbionts from a collection of marine fungi. Virus Res 219:22–38
Osaki H, Sasaki A, Nomiyama K, Tomioka K (2016) Multiple virus infection in a single strain of Fusarium poae shown by deep sequencing. Virus Genes 52:835–847
Acknowledgements
The authors thank the Platform Genotoul (INRA, Toulouse, France) for the Illumina sequencing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with animals or human participants performed by any of the authors.
Additional information
Handling Editor: Robert H.A. Coutts.
The nucleotide sequence reported in this study has been deposited in GenBank under the accession number KY906213.
Rights and permissions
About this article
Cite this article
Marais, A., Nivault, A., Faure, C. et al. Molecular characterization of a novel fusarivirus infecting the plant-pathogenic fungus Neofusicoccum luteum . Arch Virol 163, 559–562 (2018). https://doi.org/10.1007/s00705-017-3620-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3620-x