Abstract
Parainfluenza virus 5 (PIV5) is widespread in mammals and humans. Up to now, there is little information about PIV5 infection in lesser pandas. In this study, a PIV5 variant (named ZJQ-221) was isolated from a lesser panda with respiratory disease in Guangzhou zoo in Guangdong province, southern China. The full-length genome of ZJQ-221 was found to be 15,246 nucleotides and consisted of seven non-overlapping genes encoding eight proteins (i.e., NP, V, P, M, F, SH, HN and L). Sequence alignment and genetic analysis revealed that ZJQ-221 shared a close relationship with a PIV5 strain of canine-origin (1168-1) from South Korea. The findings of this study confirm the presence of PIV5 in lesser panda and indicate this mammal as a possible natural reservoir. Furthermore they highlight the urgent need to strengthen viral surveillance and control of PIV5 in zoo animals.
Similar content being viewed by others
Introduction
Members of the Paramyxoviridae family are the causative agents of mumps and measles in humans, Newcastle disease in poultry, peste des petits ruminants in sheep and goats, as well as distemper in carnivorous animals [1–4]. Parainfluenza virus 5 (PIV5) belongs to the genus Rubulavirus in the family Paramyxoviridae (http://www.ictvonline.org/virusTaxonomy.asp?taxnode_id=20151063), and was originally known as simian virus 5 (SV5) or canine parainfluenza virus (CPIV) [5–7]. PIV5 has a negative sense, single-stranded RNA genome of 15,246 nucleotides with a virion diameter of 50–200 nm that appears circular or polymorphous under the electron microscope [8–10]. PIV5 has eight viral proteins (i.e., NP, V, P, M, F, SH, HN and L). The V and P proteins are encoded by the V/P gene sharing the same genomic region [8]. PIV5 has a close genetic relationship with other members of genus Rubulavirus, including simian virus 41, human parainfluenza virus 2, human parainfluenza virus 4, mumps virus, mapuera virus and porcine rubulavirus [11–16].
Recently, paramyxoviruses have been found to have a global presence and to be prevalent in several countries [17–19]. Novel paramyxoviruses that can spread among different species have resulted in outbreaks in animals [20, 21]. Hosts susceptible to PIV5 include humans, pigs, dogs, cattle, cats, hamsters and guinea pigs [22–25]. PIV5 can cause central nervous and respiratory diseases in infected hosts [25, 26].
Up to now, there is little information about PIV5 in lesser pandas. In this study, a novel variant of PIV5 (designated as ZJQ-221) was isolated from a lesser panda with respiratory disease in Guangzhou zoo in Guangdong province, southern China. Furthermore, the complete genome sequence of ZJQ-221 was amplified and characterized.
Provenance of virus materials
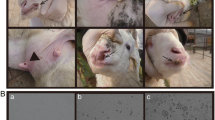
During the years of 2014–2016, fourteen representative clinical samples from ornamental animals (including four lesser pandas, one northeast tiger, one south China tiger, four Panthera leo and four ring-tailed lemurs) were collected from Guangzhou zoo in Guangdong province, southern China. Clinical manifestations of the animals included coughing with thin nasal fluid and a slightly elevated temperature. One lesser panda died, and its necropsy identified the presence of lobular pneumonia in the lungs. Nasal swabs and lung samples were stored at −80 °C until use.
Fourteen samples were tested for the possible presence of three respiratory-related pathogens (including PIV5, canine distemper virus, and coronavirus) by RT-PCR according to previous studies [27, 28]. The results revealed the positive presence of PIV5 nucleotides in the lesser panda samples. The lung samples from the lesser panda were then homogenized in 10% (w/v) sterile phosphate-buffered saline (PBS, pH 7.4), and centrifuged at 10,000 × g for 5 min at 4 °C. The supernatants were filtered aseptically (0.22 μm pore size) and the filtrates (500 μl) were inoculated onto Vero cell monolayers in a 25-cm2 cell culture flask. After viral attachment for 2 h at 37 °C, unattached viruses were removed by gentle washing with PBS and the cells were maintained in Dulbecco’s minimal essential medium (DMEM, Gibco), containing 2% fetal bovine serum (FBS, Gibco) and 1% antibiotic-antimitotic, at 37 °C in a 5% CO2 atmosphere. The viral cultures were harvested when the cytopathic effects (CPE) reached 80% coverage and then stored at −80 °C [25]. In order to observe the virus particles, the cell suspensions were centrifuged at 1,000 rpm for 10 min, and then fixed using 2.5 ~ 3% glutaraldehyde fixation fluid for about 4 h at 4 °C in a refrigerator. Finally, they were submitted to the electron microscope center, South China Agricultural University for processing.
RNA extraction of positive homogenates was performed using the MiniBEST Universal RNA Extraction Kit (TaKaRa), and viral RNA was reverse transcribed into first-strand cDNA with a random primer (5′-NNNNNN-3′) using the PrimeScript first strand cDNA Synthesis Kit (TaKaRa). PCR was then used to amplify the full-length genome sequences using 12 pairs of primers listed in Table S1. Positive PCR products were purified (MiniBEST Agarose Gel DNA Extraction Kit, TaKaRa) and cloned into the pMD19-T vector (TaKaRa). Positive recombinant plasmids were further sequenced using the Sanger sequencing method (BGI Inc., Guangzhou branch). Sequence alignment and phylogenetic analysis based on different rubulavirus species sequences (Table 1) was performed using DNAStar Lasergene 7.10 (Madison, WI, USA) and MEGA 5 (http://www.megasoftware.net) [34].
Sequence properties
In comparison with uninfected cells (Fig. S1a), Vero cells inoculated with PIV5-positive samples showed demonstrable CPE by passage 3 (Fig. S1b). One viral strain (named ZJQ-221) was thus obtained. Further observation by electron microscopy found that the isolated ZJQ-221 strain was spherical and had a diameter of 50-200 nm that is similar to, and characteristic of, paramyxoviruses (Fig. S1c). Moreover, through sequencing, the full-length genome sequence (15,246 nucleotides, nt) of ZJQ-221 was obtained, which included a 3′ leader sequence (55 nt), seven non-overlapping encoding sequences (i.e., NP gene at positions 152-1681, V/P gene at positions 1850-2518, M gene at positions 3141-4274, F gene at positions 4530-6185, SH gene at positions 6303-6437, HN gene at positions 6584-8281 and L gene at positions 8414-15181, respectively) and a 5′ trailer sequence (31 nt). Complete genome sequence alignments showed that ZJQ-221 had the highest nucleotide similarity (~ 99.8%) with a PIV5 strain of canine-origin (1168-1, GenBank accession no. KC237064) from South Korea and the lowest nucleotide similarity (~ 97.2%) with a PIV5 strain of canine-origin (D277, GenBank accession no. KC237065), respectively (Table S2). Between 1168-1 and ZJQ-221, there were 3-nt, 4-nt, 2-nt, 4-nt, 5-nt, 10-nt and 1-nt differences in the 3′ leader, NP, M, F, HN, L and 5′ trailer, respectively. At the amino acid level, they differed at the following positions: NP (Glu → Gly at position 108), M (Glu → Ala at position 375), F (Val → Ala, Thr → Ala, Gln → Arg and Asp → Ala at positions 134, 279, 339 and 445, respectively), HN (Leu → Ser, Phe → Leu, and Asn → Thr at positions 15, 210 and 288, respectively), and L (Arg → Lys and Arg → Leu at positions 1631 and 2248). In addition, the SH protein was not present in some PIV5 strains due to nucleotide substitution, such as KUN-11 (KC852177), SER (JQ743328), CC14 (KP893891) PV5-BC14 (KM067467) and AGS (KX060176), implying that the protein is not essential for PIV5 infection in pigs, dogs, calves and cells [25, 29, 30, 32, 33]. However, the ZJQ-221 strain had a SH protein and was thus different from the above-mentioned PIV5 strains. Furthermore, phylogenetic analysis (Fig. 1) was performed based on the complete genome sequences of different species of rubulaviruses. In the phylogenetic tree, ZJQ-221 and 1168-1 shared the closest genetic relationship and were clustered in the same branch (Fig. 1).
Phylogenetic analysis of ZJQ-221 and other rubulaviruses. A phylogenetic tree based on the complete genome sequences of different rubulavirus species was constructed using the Maximum-likelihood method. ZJQ-221 is displayed using a red solid box. Different rubulavirus species within the Rubulavirus genus are marked using blue solid boxes. The origins of the different viral strains or isolates are indicated with different symbols
To the best of our knowledge, very little data about PIV5 infections in zoo animals is available. In this study, we tested whether PIV5 infections are present in lesser pandas, northeast tigers, south China tigers, Panthera leo and ring-tailed lemurs. While the RT-PCR results showed that only lesser panda samples were positive for PIV5, due to the limited number of animal samples collected in the zoo, we believe that PIV5 might not be restricted to lesser pandas. In fact, a previous study showed antibodies against PIV5 exist in zoo tigers [35]. Moreover, PIV5 nucleotide sequences were also detected in nasal swab samples from northeast tiger and south China tiger collected in 2016 in the same zoo, and their F genes were close to the lesser panda-origin PIV5 strain described here (data not shown). This suggests that PIV5 is a common pathogen in zoo animals, and might play an important causal role in the respiratory diseases of zoo animals.
In summary, we have identified a novel PIV5 isolate in lesser panda and performed whole genome sequencing, indicating that this mammal may act as a possible natural reservoir for this virus. This study contributes to the epidemiology and genomics of PIV5, and suggests an urgent need to strengthen viral surveillance and control of PIV5 in zoo animals.
Nucleotide sequence accession number: The complete genome sequence of the ZJQ-221 isolate has been deposited in GenBank under the accession number KX100034.
References
Bale JF Jr (2014) Measles, mumps, rubella, and human parvovirus B19 infections and neurologic disease. Handb Clin Neurol 121:1345–1353
Dimitrov KM, Ramey AM, Qiu X, Bahl J, Afonso CL (2016) Temporal, geographic, and host distribution of avian paramyxovirus 1 (Newcastle disease virus). Infect Genet Evol 39:22–34
Li XP, Zhai SL, He DS, Guo PJ, Lv DH, Wen XH, Luo ML, Chen RA, Wei WK (2015) Genome characterization and phylogenetic analysis of a lineage IV peste des petits ruminants virus in southern China. Virus Genes 51:361–366
Martinez-Gutierrez M, Ruiz-Saenz J (2016) Diversity of susceptible hosts in canine distemper virus infection: a systematic review and data synthesis. BMC Vet Res 12:78
Hull RN, Minner JR, Smith JW (1956) New viral agents recovered from tissue cultures of monkey cells. I. Origin and properties of cytopathogenic agents SV1, SV2, SV4, SV5, SV6, SV11, SV12 and SV15. Am J Hyg 63:204–215
Choppin PW (1964) Multiplication of a myxovirus (SV5) with minimal cytopathic effects and without interference. Virology 23:224–233
Evermann JF, Krakowka S, McKeirnan AJ, Baumgärtner W (1981) Properties of an encephalitogenic canine parainfluenza virus. Arch Virol 68:165–172
He B, Paterson RG, Ward CD, Lamb RA (1997) Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology 237:249–260
Choppin PW, Stoeckenius W (1964) The morphology of SV5 virus. Virology 23:195–202
Terrier O, Rolland JP, Rosa-Calatrava M, Lina B, Thomas D, Moules V (2009) Parainfluenza virus type 5 (PIV-5) morphology revealed by cryo-electron microscopy. Virus Res 142:200–203
Ogawa M, Mutsuga N, Tsurudome M, Kawano M, Matsumura H, Kusagawa S, Komada H, Nishio M, Ito Y (1992) Nucleotide sequence analysis of the simian virus 41 gene encoding the large (l) protein and construction of a phylogenetic tree for the l proteins of paramyxoviruses. J Gen Virol 73:2743–2750
Skiadopoulos MH, Vogel L, Riggs JM, Surman SR, Collins PL, Murphy BR (2003) The genome length of human parainfluenza virus type 2 follows the rule of six, and recombinant viruses recovered from non-polyhexameric-length antigenomic cDNAs contain a biased distribution of correcting mutations. J Virol 77:270–279
Komada H, Kawano M, Uefuji A, Ito M, Tsurudome M, Hatakeyama E, Nakanishi M, Sakue S, Joh C, Suzumura E, Tamaki T, Tomioka T, Nishio M, Tsumura H, Uematsu J, Yamamoto H, O’Brien M, Bando H, Ito Y (2011) Completion of the full-length genome sequence of human parainfluenza virus types 4A and 4B: sequence analysis of the large protein genes and gene start, intergenic and end sequences. Arch Virol 156:161–166
Lee JY, Na BK, Lee HD, Chang SW, Kim KA, Kim JH, Cho HW, Kim J, Kang C (2004) Complete nucleotide sequence of a mumps virus genotype I strain isolated in Korea. Virus Genes. 28(2):201–205
Tillieux SL, Halsey WS, Sathe GM, Vassilev V (2009) Comparative analysis of the complete nucleotide sequences of measles, mumps, and rubella strain genomes contained in Priorix-Tetra and ProQuad live attenuated combined vaccines. Vaccine 27:2265–2273
Wang LF, Hansson E, Yu M, Chua KB, Mathe N, Crameri G, Rima BK, Moreno-López J, Eaton BT (2007) Full-length genome sequence and genetic relationship of two paramyxoviruses isolated from bat and pigs in the Americas. Arch Virol 152:1259–1271
Nagao Y, Nishio Y, Shiomoda H, Tamaru S, Shimojima M, Goto M, Une Y, Sato A, Ikebe Y, Maeda K (2012) An outbreak of canine distemper virus in tigers (Panthera tigris): possible transmission from wild animals to zoo animals. J Vet Med Sci 74:699–705
Cêtre-Sossah C, Kwiatek O, Faharoudine A, Soulé M, Moutroifi YO, Vrel MA, Salami H, Rassoul S, Asnaoui M, Moindjie Y, Albina E, Libeau G, Cardinale E (2016) Impact and Epidemiological Investigations into the Incursion and Spread of Peste des Petits Ruminants in the Comoros Archipelago: an increased threat to surrounding Islands. Transbound Emerg Dis 63:452–459
Chua KB, Bellini WJ, Rota PA, Harcourt BH, Tamin A, Lam SK, Ksiazek TG, Rollin PE, Zaki SR, Shieh W, Goldsmith CS, Gubler DJ, Roehrig JT, Eaton B, Gould AR, Olson J, Field H, Daniels P, Ling AE, Peters CJ, Anderson LJ, Mahy BW (2000) Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432–1435
Rentería-Solís Z, Förster C, Aue A, Wittstatt U, Wibbelt G, König M (2014) Canine distemper outbreak in raccoons suggests pathogen interspecies transmission amongst alien and native carnivores in urban areas from Germany. Vet Microbiol 174:50–59
Field HE, Breed AC, Shield J, Hedlefs RM, Pittard K, Pott B, Summers PM (2007) Epidemiological perspectives on Hendra virus infection in horses and flying foxes. Aust Vet J 85:268–270
Chatziandreou N, Stock N, Young D, Andrejeva J, Hagmaier K, McGeoch DJ, Randall RE (2004) Relationships and host range of human, canine, simian and porcine isolates of simian virus 5 (parainfluenza virus 5). J Gen Virol 85:3007–3016
Hsiung GD (1972) Parainfluenza-5 virus. Infection of man and animal. Prog Med Virol 14:241–274
Yang DK, Nah JJ, Kim HH, Choi SS, Bae YC, Park JW, Song JY (2014) Isolation of novel bovine parainfluenza virus type 5 (bPIV5) and its incidence in Korean cattle. Korean J Vet Res 54:107–112
Liu Y, Li N, Zhang S, Zhang F, Lian H, Hu R (2015) Parainfluenza Virus 5 as possible cause of severe respiratory disease in Calves, China. Emerg Infect Dis 21:2242–2244
Baumgärtner WK, Krakowka S, Koestner A, Evermann J (1982) Acute encephalitis and hydrocephalus in dogs caused by canine parainfluenza virus. Vet Pathol 19:79–92
Romanutti C, Gallo Calderón M, Keller L, Mattion N, La Torre J (2016) RT-PCR and sequence analysis of the full-length fusion protein of Canine Distemper Virus from domestic dogs. J Virol Methods 228:79–83
Decaro N, Mari V, Larocca V, Losurdo M, Lanave G, Lucente MS, Corrente M, Catella C, Bo S, Elia G, Torre G, Grandolfo E, Martella V, Buonavoglia C (2016) Molecular surveillance of traditional and emerging pathogens associated with canine infectious respiratory disease. Vet Microbiol 192:21–25
Rima BK, Gatherer D, Young DF, Norsted H, Randall RE, Davison AJ (2014) Stability of the parainfluenza virus 5 genome revealed by deep sequencing of strains isolated from different hosts and following passage in cell culture. J Virol 88:3826–3836
Gao S, Jin H, Zhang S, Liu Y, Hu R (2015) The complete genome sequencing and analysis of canine parainfluenza virus strain CC-14. Chin J Prev Vet Med 37:802–804 (in Chinese)
Oem JK, Kim SH, Kim YH, Lee MH, Lee KK (2015) Molecular characteristics of canine parainfluenza viruses type 5 (CPIV-5) isolated in Korea. Can J Vet Res 79:64–67
Lee YN, Lee C (2013) Complete genome sequence of a novel porcine parainfluenza virus 5 isolate in Korea. Arch Virol 158:1765–1772
Wignall-Fleming E, Young DF, Goodbourn S, Davison AJ, Randall RE (2016) Genome sequence of the parainfluenza virus 5 strain that persistently infects AGS cells. Genome Announc 4:e00653-16
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Sun H, Xia X, Gao Y, He W, Wang L, Liu Huang G (2004) Seroepidemiological investigation of caine parinfluenza virus in tiger. Anim Husb Vet Med 36:4–5 (In Chinese)
Acknowledgements
This work was mostly supported by Guangzhou Zoo. Moreover, this work was also partially supported by Ministry of Science and Technology of the People’s Republic of China (Grant No. 2015GA780010), Guangdong Provincial Department of Science and Technology (Grant Nos. 2016A040403083 and 2016B020234006), Guangdong Provincial Agricultural Department (Grant No. 2016LM3177), and Guangzhou Science Technology and Innovation Commission (Grant No. 201508020055).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Jun-Qiong Zhai, Shao-Lun Zhai and Tao Lin contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2017_3245_MOESM3_ESM.pdf
Figure S1 In vitro proliferation and viral particle characteristics of the ZJQ-221 strain. (a) Normal Vero cells (40 ×); (b) Vero cells infected with the ZJQ-221 strain (40 ×); (c) Viral particles of ZJQ-221 were observed under the electron microscope (×30, 000) (PDF 97 kb)
Rights and permissions
About this article
Cite this article
Zhai, JQ., Zhai, SL., Lin, T. et al. First complete genome sequence of parainfluenza virus 5 isolated from lesser panda. Arch Virol 162, 1413–1418 (2017). https://doi.org/10.1007/s00705-017-3245-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3245-0