Abstract
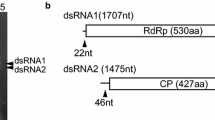
Plasmopara halstedii virus (PhV) is one of the few characterized oomycete viruses. Although it is fully sequenced and well-studied in its genetic diversity, the exact classification and phylogenetic relationships of PhV remain uncertain. The only known virus with characteristics similar to PhV is the Sclerophthora macrospora Virus A (SmV-A). Both viruses infect obligate biotrophic oomycetes. While RNA-dependent RNA polymerases (RdRp) of oomycetes viruses have high similarity to the corresponding enzymes from viruses classified in the family Nodaviridae, the coat proteins (CP) seem to be completely different from those of other viruses of this family. In contrast, the coat proteins of PhV and SmV-A have high similarity to viruses classified in the Tombusviridae, Circoviridae and a new group of hybrid DNA-RNA viruses (so-called chimeric viruses or cruciviruses). Because phylogenetic analyses based on the sequences of either RdRp or CP result in different affinities, an alternative, genome-based approach combining the sequences of both proteins was used. This analysis placed the two oomycete viruses together with Tombunodavirus UC1 in a new, independent group between families Nodaviridae and Tombusviridae.



Similar content being viewed by others
References
Heller-Dohmen M, Göpfert JC, Pfannenstiel J, Spring O (2011) The nucleotide sequence and genome organization of Plasmopara halstedii virus. Virol J 8:123–131
Yokoi T, Takemoto Y, Suzuki M, Yamashita S, Hibi T (1999) The nucleotide sequence and genome organization of Sclerophthora macrospora Virus B. Virology 264:344–349
Yokoi T, Yamashita S, Hibi T (2003) The nucleotide sequence and genome organization of Sclerophthora macrospora Virus A. Virology 311:394–399
Gulya TJ, Freeman TP, Mayhew DE (1992) Ultrastructure of virus-like particles in Plasmopara halstedii. Can J Bot 70:334–339
Heller-Dohmen M, Göpfert JC, Hammerschmidt R, Spring O (2008) Different pathotypes of the sunflower downy mildew pathogen Plasmopara halstedii all contain isometric virions. Mol Plant Pathol 9:777–786
Grasse W, Spring O (2015) Occurrence and genetic diversity of Plasmopara halstedii virus in sunflower downey mildew populations of the world. Fungal Biol 119:170–178
Grasse W, Zipper R, Totska M, Spring O (2013) Plasmopara halstedii virus causes hypovirulence in Plasmopara halstedii, the downy mildew pathogen of the sunflower. Fungal Genet Biol 57:42–47
Cai G, Myers K, Hillman BI, Fry WE (2009) A novel virus of the late blight pathogen, Phytophthora infestans, with two RNA segments and a supergroup 1 RNA-dependent RNA polymerase. Virology 392:52–61
Cai G, Myers K, Fry WE, Hillman BI (2012) A member of the virus family Narnaviridae from the plant pathogenic oomycete Phytophthora infestans. Arch Virol 157:165–169
Cai G, Krychiw JF, Myers K, Fry WE, Hillman BI (2013) A new virus from the plant pathogenic oomycete Phytophthora infestans with an 8 kb ds RNA genome: The sixth member of a proposed new virus genus. Virology 435:341–349
Diemer GS, Stedman KM (2012) A novel virus genome discovered in an extreme environment suggests recombination between unrelated groups of RNA and DNA viruses. Biol Direct 7:13
Roux S, Enault F, Bronner G, Vaulot D, Forterre P, Kropovic M (2013) Chimeric viruses blur the borders between the major groups of eukaryotic single-stranded DNA viruses. Nature Communications 4:2700
Krupovic M, Zhi N, Li J, Hu G, Koonin EV, Wong S, Shevchenko S, Zhao K, Young NS (2015) Multiple layers of chimerism in a single-stranded DNA virus discovered by deep sequencing. Genome Biol Evol 7(4):993–1001
Ahola T, Karlin DG (2015) Sequence analyses reveals a conserved extension in the capping enzyme of the alphavirus soupergroup, and homologous domain in nodaviruses. Biol Direct 10:16
Runckel C, Flenniken ML, Engel JC, Ruby JG, Ganem D, Andino R, deRisi JL (2011) Temporal analysis of the honey bee microbiome reveals four novel viruses and seasonal prevalence of known viruses, Nosema, and Crithidia. PLoS One 6(6):e20656
Greninger AL, deRisi JL (2015) Draft genome sequence of Tombunodavirus UC1. Genome Announcements 3(4):e00655-15
Greninger AL, Kellogg M, DeRisi JL (2013) (submitted, 2013 unpublished) Virus sequences identified in SF wastewater. Biochemistry, UCSF
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Molec Biol Evol 30:2725–2729
Stedman K (2013) Mechanisms for RNA capture by ssDNA viruses: grand theft RNA. J Molec Evol 76(6):359–364
Acknowledgements
We gratefully acknowledge the valuable comments and assistance of Margaret Janke, Institut of Botany, University of Hohenheim, for help preparing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Grasse, W., Spring, O. ssRNA viruses from biotrophic Oomycetes form a new phylogenetic group between Nodaviridae and Tombusviridae . Arch Virol 162, 1319–1324 (2017). https://doi.org/10.1007/s00705-017-3243-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3243-2