Abstract
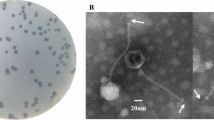
A novel virulent enterobacteria phage, 4MG, which was isolated from soil near a sewer, belongs to the family Myoviridae, as it possesses an isometric head and a long contractile tail. The complete genome of 4MG consists of a double-stranded DNA with a length of 148,567 bp, a G + C content of 46.3 %, 271 open reading frames (ORFs), and 21 tRNAs. Bioinformatic analysis revealed that 4MG highly resembles “rV5-like viruses” but can be separated, together with Salmonella phage PVP-SE1 and Cronobacter sakazakii phage vB_CsaM_GAP31, as part of the subgroup “PVP-SE1-like phage”.


Similar content being viewed by others
References
Lavigne R, Darius P, Summer EJ, Seto D, Mahadevan P, Nilsson AS, Ackermann HW, Kropinski AM (2009) Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol 9:224
Santos SB, Kropinski AM, Ceyssens PJ, Ackermann HW, Villegas A, Lavigne R, Krylov VN, Carvalho CM, Ferreira EC, Azeredo J (2011) Genomic and proteomic characterization of the broad-host-range Salmonella phage PVP-SE1: creation of a new phage genus. J Virol 85:11265–11273
Kropinski AM, Waddell T, Meng J, Franklin K, Ackermann HW, Ahmed R, Mazzocco A, Yates J 3rd, Lingohr EJ, Johnson RP (2013) The host-range, genomics and proteomics of Escherichia coli O157:H7 bacteriophage rV5. Virol J 10:76
Abbasifar R, Kropinski AM, Sabour PM, Ackermann HW, Alanis Villa A, Abbasifar A, Griffiths MW (2012) Genome sequence of Cronobacter sakazakii myovirus vB_CsaM_GAP31. J Virol 86:13830–13831
Schwarzer D, Buettner FF, Browning C, Nazarov S, Rabsch W, Bethe A, Oberbeck A, Bowman VD, Stummeyer K, Mühlenhoff M, Leiman PG, Gerardy-Schahn R (2012) A multivalent adsorption apparatus explains the broad host range of phage phi92: a comprehensive genomic and structural analysis. J Virol 86:10384–10398
Truncaite L, Šimoliūnas E, Zajančkauskaite A, Kaliniene L, Mankevičiūte R, Staniulis J, Klausa V, Meškys R (2012) Bacteriophage vB_EcoM_FV3: a new member of “rV5-like viruses”. Arch Virol 157:2431–2435
Tsonos J, Adriaenssens EM, Klumpp J, Hernalsteens JP, Lavigne R, De Greve H (2012) Complete genome sequence of the novel Escherichia coli phage phAPEC8. J Virol 86:13117–13118
Kim M, Ryu S (2011) Characterization of a T5-like coliphage, SPC35, and differential development of resistance to SPC35 in Salmonella enterica serovar Typhimurium and Escherichia coli. Appl Environ Microbiol 77:2042–2050
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Besemer J, Lomsadze A, Borodovsky M (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618
Delcher AL, Bratke KA, Powers EC, Salzberg SL (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Zdobnov EM, Apweiler R (2001) InterProScan––an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848
Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, Ke Z, Krylov D, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Thanki N, Yamashita RA, Yin JJ, Zhang D, Bryant SH (2007) CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res 35:D237–D240
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964
Sullivan MJ, Petty NK, Beatson SA (2011) Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010
Mahadevan P, King JF, Seto D (2009) CGUG: in silico proteome and genome parsing tool for the determination of “core” and unique genes in the analysis of genomes up to ca. 1.9 Mb. BMC Res Notes 2:168
Kim M, Ryu S (2012) Spontaneous and transient defence against bacteriophage by phase-variable glucosylation of O-antigen in Salmonella enterica serovar Typhimurium. Mol Microbiol 86:411–425
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Acknowledgments
This work was carried out with the support of “Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ009842)”, Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kim, M., Heu, S. & Ryu, S. Complete genome sequence of enterobacteria phage 4MG, a new member of the subgroup “PVP-SE1-like phage” of the “rV5-like viruses”. Arch Virol 159, 3137–3140 (2014). https://doi.org/10.1007/s00705-014-2140-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-014-2140-1