Abstract
Cynomolgus macaques are widely used as an animal model in biomedical research. We have established an immortalized cynomolgus macaque fibroblast cell line (MSF-T) by transducing primary dermal fibroblasts isolated from a 13-year-old male cynomolgus macaque with a retrovirus vector expressing human telomerase reverse transcriptase (hTERT). The MSF-T cells showed increased telomerase enzyme activity and reached over 200 in vitro passages compared to the non-transduced dermal fibroblasts, which reached senescence after 43 passages. The MSF-T cell line is free of mycoplasma contamination and is permissive to the newly identified cynomolgus macaque cytomegalovirus (CyCMV). CyCMV productively infects MSF-T cells and induces down-regulation of MHC class I expression. The MSF-T cell line will be extremely useful for the propagation of CyCMV and other cynomolgus herspesviruses in host-derived fibroblast cells, allowing for the retention of host-specific viral genes. Moreover, this cell line will be beneficial for many in vitro experiments related to this animal model.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cynomolgus macaques (Macaca fascicularis) are a routinely used non-human primate (NHP) model to study genetic diseases, physiology, behaviour, neuroscience, metabolic and infectious diseases in humans. Recently, cynomolgus macaques have become one of the most popular animal models among HIV vaccine researchers [4, 33]. Despite their broad use, few cell lines are available for in vitro studies related to this animal model [2, 9, 12, 37, 39, 40]. Cultured fibroblasts are particularly useful in medical research as they provide a useful model for conducting experiments in a controlled environment. However, in vitro, primary fibroblasts change considerably as they undergo replication and have a finite replicative lifespan. Maintaining short-term cultures of primary fibroblasts can alleviate this problem; however, it is labour intensive, and in addition, cells from different animals may contain adventitious agents that can influence the reproducibility of the experiments. This can be circumvented by immortalization of primary fibroblasts, which can be achieved by a number of methods including chemical treatments, viral oncogene expression and constitutive ectopic expression of human telomerase reverse transcriptase (hTERT).
Cytomegaloviruses are highly species-specific, ubiquitous herpesviruses that cause asymptomatic lifelong infections in immunocompetent individuals but life-threatening infections in immunocompromised individuals and in newborns. Rhesus macaque CMV (RhCMV) is the most widely used non-human primate model for human CMV (HCMV) research [44]. Recently, we have isolated and characterized a novel cytomegalovirus from a 4-year-old female cynomolgus macaque housed at the non-human primate colony at Health Canada, Ottawa, Canada [3]. The complete genome of this cynomolgus macaque cytomegalovirus (CyCMV) has since been sequenced and annotated using next-generation Illumina sequencing [26]. CyCMV may provide an alternative non-human primate model for understanding HCMV biology and pathogenesis and for vaccine development. Furthermore, CyCMV offers another model to evaluate CMV-based HIV vaccines. CyCMV (Ottawa strain) was isolated by inoculating commercially available human diploid lung fibroblast cells (MRC-5) with catheterized urine samples from a cynomolgus macaque. MRC-5 cells were chosen because there was no cynomolgus fibroblast cell line available. A number of non-human primate cytomegaloviruses have been isolated and propagated in MRC-5 cells; however, studies with RhCMV have shown that RhCMV grows better in a host-specific telomerase-transduced rhesus fibroblast (Telo-RF) cell line [8].
In order to facilitate CyCMV-related research and other in vitro studies that require a continuous supply of cynomolgus macaque fibroblasts, we have established and characterized an immortal cynomolgus macaque fibroblast cell line by retroviral-mediated ectopic expression of hTERT. The cell line shows constitutive expression of hTERT and telomerase activity, exhibits stable growth kinetics and supports the growth of CyCMV.
Materials and methods
Cells and viruses
MRC-5 cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA) and used within 5-25 serial passages. Telo-RF cells were kindly provided by Dr. P. Barry at the University of California-Davis, CA [23]. Both cell types were maintained in DMEM (Invitrogen) supplemented with 10 % FBS (Wisent Bioproducts, Quebec), 100 U/ml penicillin and 100 μg/ml streptomycin (Sigma). CyCMV (Ottawa strain) was propagated and titered on MRC-5 cells as described previously [3].
Isolation of cynomolgus macaque dermal fibroblasts
Six punch biopsies (4-6 mm in diameter) were collected from the lateral thigh region of a 13-year-old male monkey (Mellow-C95073 M) that was housed at the non-human primate facility at Health Canada, Ottawa, Canada. The biopsies were immediately placed in Hank’s balanced salt solution (HBSS) supplemented with 10 % FBS, penicillin (100 IU/ml), streptomycin (100 μg/ml), amphotericin B (0.25 μg/ml), and shipped overnight on ice to the University of Toronto, Canada. Immediately upon arrival, the biopsies were transferred to a Petri dish containing 10 ml of 0.25 % trypsin in PBS and incubated at room temperature for 30 min. The dermis was isolated from the epidermis of each biopsy using sterile forceps, cut into small pieces and transferred to an Eppendorf tube containing 1 ml of an enzyme cocktail (1 mM sodium pyruvate, 2.73 mg/ml bacterial collagenase, 1.25 mg/ml hyaluronidase, 0.1 mg/ml DNase I in RPMI supplemented with HEPES). The tubes were agitated at 37 °C in a shaking incubator for approximately 1-2 h until no tissue clumps were visible. The cell suspensions were pooled and filtered through a sterile 100-μm nylon mesh (BD Biosciences) to remove any remaining tissue fragments. The filtrate was centrifuged at 1200 rpm for 10 min at room temperature. The resulting cell pellet was resuspended in 12 ml culture medium (DMEM supplemented with 10 % FBS, penicillin [100 IU/ml], streptomycin [100 μg/ml] and amphotericin B [0.25 μg/ml]), transferred into four wells of a 6-well tissue culture plate and incubated at 37 °C overnight in a 5 % CO2 incubator. Twenty-four hours later, non-adherent cells were removed, fresh culture media was added, and the plate was incubated until the primary cells formed a confluent monolayer. The cells were trypsinized and split at a 1:2 ratio until sufficient numbers of fibroblasts were obtained. Multiple vials of fibroblasts (at passage #8) were frozen in liquid nitrogen, and the primary fibroblasts were termed Mellow (name of the macaque from which they were derived) skin fibroblast (MSF) cells.
Pseudotyping Moloney retrovirus with VSV G protein
The retroviral vector pLXIN (Stratagene) expressing hTERT was kindly provided by Dr. G.S. Pari at the University of Nevada-Reno, Nevada, USA. The plasmid pME-VSV-G encoding vesicular stomatitis virus (VSV) glycoprotein was obtained from Dr. Tania Watts at the University of Toronto, ON, Canada. The day prior to transfection, Phoenix cells (second-generation retrovirus packaging cell line created by the Nolan laboratory at Stanford University) were plated at 2.5 × 106 cells per 60-mm plate in DMEM supplemented with 10 % FBS. Just before transfection, cell culture medium was removed, fresh culture medium containing 25 μM chloroquine was added, and the plate was returned to the incubator. Using the calcium phosphate method, cells were co-transfected with 5 μg of pME-VSV-G and 5 μg of pLXIN-hTERT. Approximately 10 h later, the culture medium was removed and replaced with 5 ml of fresh cell culture medium. Retroviral supernatant was harvested every 12 h starting 24 h post-transfection until 72 h post-transfection. The supernatant was filtered through a 45-μm syringe filter to remove any live cells and concentrated by ultracentrifugation at 25,000 rpm for 90 min at 4 °C.
Transduction and selection of primary cynomolgus fibroblasts
Primary cynomolgus dermal fibroblast (MSF) cells at passage #8 were subjected to different concentrations of Geneticin (G418, Invitrogen) to determine the minimum G418 concentration required to abolish the cell monolayer. Fibroblasts (passage #8) were grown to confluency in a 60-mm tissue culture dish and infected with concentrated VSV G-pseudotyped hTERT expressing Moloney retroviruses in growth medium containing 8 μg/ml Polybrene (Santa Cruz Biotechnology, Inc.) for 3 h at 37 °C. The Polybrene concentration was reduced to 4 μg/ml by adding fresh growth medium without Polybrene, and the cells were maintained at 37 °C in a 5 % CO2 incubator. Forty-eight hours after the transduction, the inoculum was removed, and the transduced cells were selected with 150 μg/ml G418. The cell clones that survived the G418 selection over 3-4 weeks were expanded and maintained in growth medium supplemented with 100 μg/ml G418.
Telomerase RT-PCR assay
Total RNA from 2.5 × 106 Telo-RF, MSF and MSF-T cells was isolated using an RNeasy Mini Kit (QIAGEN). cDNA was synthesized from 1.8 μg of total RNA from each cell type using Omniscript Reverse Transcriptase (QIAGEN) and oligo-dT primers according to the manufacturer’s instructions. cDNA was amplified with primers specific for hTERT (FP, 5′CGGAAGAGTGTCTGGAGCAA3′, and RP, 5′GGATG AAGCGGAGTCTGGA3′). Beta-actin primers (FP, 5′GTGGGGCGCCCCAGGCACCA3′ and RP, 5′CTCCTTAATGTCACGCACGATTTC 3′) were used as a positive control. PCR conditions were as follows: 94 °C for 2 min followed by 34 cycles of 94 °C for 45 sec, 53 °C for 30 sec, and 72 °C for 1 min, and a final extension at 72 °C for 1 min. PCR amplicons were resolved on a 1 % agarose gel, stained with ethidium bromide and visualized using an AlphaImager 2200 (Alpha Innotech).
Telomerase assay
Telomerase activity in MSF and MSF-T cells were compared using a TRAPeze® Telomerase Detection Kit (Chemicon) according to manufacturer’s instructions. Briefly, 2.5 × 106 MSF and MSF-T cells were lysed in 200 μl of cold lysis buffer for 30 min on ice, and cellular debris was spun down at 15,000 rpm for 20 min at 4 °C. The cell extract (2 μl of each) was incubated with 48 μl of 10× TRAP reaction buffer supplemented with dNTP, TS primers, TRAP primer mix, and Taq DNA polymerase for 30 min at 30 °C. The telomerase-extended TS primers were amplified by PCR and visualized on a 12 % non-denaturing polyacrylamide gel (Invitrogen).
Mycoplasma detection
MSF-T cells at 75 population doublings (PD) were tested for mycoplasma contamination using a VenorGeM® Mycoplasma Detection Kit (Sigma) according to the manufacturer’s instructions. To avert false negative reactions, MSF-T cells were grown in antibiotic- free culture medium for 7-10 days prior to mycoplasma detection. Briefly, 100 μl of cell culture supernatant was heated at 95 °C for 5 min and centrifuged to pellet any cell debris, and 2 μl of the clarified supernatant was used for PCR. The VenorGeM® Mycoplasma Detection Kit detects all Mycoplasma, Acholeplasma, and Ureaplasma species, which are usually encountered as contaminants in cell cultures.
CyCMV replication kinetics
To compare the replication kinetics of CyCMV (Ottawa strain) in MSF-T, Telo-RF and MRC-5 cells, the cells were grown in 6-well tissue culture plates. The confluent monolayers were inoculated (in duplicates) with 500 μl of CyCMV at two different multiplicities of infections (0.3 and 0.03). After 2 h of adsorption at 37 °C, 1 ml of fresh cell culture medium was added to each well. The cells and the culture supernatant were harvested on day 2, 4, 6, 8 and 10 post-infection and frozen at −80 °C with 0.2 M sucrose added. The viral titers in each sample were determined by standard plaque assay on MRC-5 cells. Briefly, tenfold serial dilutions of each sample were prepared and used to inoculate confluent monolayers of MRC-5 cells in 6-well plates. After 2 h of adsorption at 37 °C, the inoculum was removed and the cell monolayer was overlaid with a 1:1 mixture of 2× MEM (containing 10 % FBS and 4 mM L-glutamine; Quality Biologicals Inc., Maryland, USA) and 2 % low-melting agarose (type VII; Sigma). The plates were incubated at 37 °C for 12-14 days, and the plaques at the end-point dilutions were counted.
Flow cytometry and CFSE labeling
For the detection of MHC class I expression, mock- or CyCMV-infected MSF-T, Telo-RF and MRC-5 cells were trypsinized and immediately resuspended in FACS buffer (PBS + 3 % FBS + 0.1 % sodium azide). The cells were incubated with PE-conjugated W6/32 antibody (eBioscience) for 45 min at 4 °C, washed four times in ice-cold FACS buffer and analyzed on a BD FACSCalibur (BD Biosciences).
CFSE labeling of MSF and MSF-T cells was done using a CellTrace CFSEcell Proliferation Kit (Invitrogen) according to the adherent cell labeling protocol recommended by the manufacture. The samples were analyzed on a BD FACSCalibur, and the histograms were generated using the FLOWJO software (Tree Star Inc.)
Western blotting
Mock- or CyCMV-infected MSF-T, Telo-RF and MRC-5 cells were lysed in RIPA buffer (50 mM Tris HCl, pH 8, 150 mM NaCl, 1 % NP-40, 0.5 % sodium deoxycholate, 0.1 % SDS, and 1 mM EDTA, adjusted to pH 7.4). Total clarified protein (10 μg) from each sample was resolved on a 4-20 % TGX gel (Bio-Rad) and transferred to a nitrocellulose membrane (GE). The membrane was probed with HC-10 [36], rabbit anti-β2 microglobulin (M8523, Sigma) or mouse anti-actin (Sigma) antibodies, followed by the relevant secondary antibodies (anti-mouse-HRP or anti-rabbit-HRP, Cedarlane), and proteins were detected using a Luminata Western HRP Kit (Millipore) according to the manufacturer’s instructions.
Results
hTERT-extended life span of cynomolgus macaque fibroblasts
The adherent primary cynomolgus macaque dermal fibroblast (MSF) cells resulting from the enzymatic digestion of skin biopsies were expanded and cryopreserved following eight passages in culture. The MSF cells were morphologically similar to other mammalian fibroblasts, and during initial passages they reached 100 % confluency in 4-5 days following each 1:2 split. To immortalize the primary fibroblasts, MSF cells were infected with VSV-G-pseudotyped hTERT-expressing Moloney retroviruses, and the transduced cells were selected with G418. The cell clones that survived the lethal concentration of G418 (150 μg/ml, as determined by a kill curve, data not shown) were expanded, and the population doubling (PD) of one clone (MSF-T) was measured with a split ratio of 1:2 (Fig. 1a). The MSF-T clone showed a slightly decreased doubling time (3 days) compared to the MSF cells (4 days). The rates of proliferation of the two cell types were compared using CFSE labeling (Fig. 1b). The cells were labelled with CFSE for 15 min and were subsequently analyzed after 1, 2, 3 and 5 days by flow cytometry for the twofold reduction in fluorescence intensity following each cell division. The fluorescence intensity of the MSF-T cells decreased faster than that of the MSF cells, confirming that the MSF-T cells proliferate at a faster rate than the MSF cells. The decreased rate of reduction in fluorescence intensity observed between 72 h and 120 h post-labeling in MSF-T cells was due to contact inhibition of the cells in the plate after 3 days.
(a): hTERT extends the lifespan of cynomolgus macaque dermal fibroblasts. Cynomolgus macaque primary dermal fibroblasts (MSF cells) at passage # 8 were transduced with VSV-G-pseudotyped hTERT expressing Moloney retrovirus and selected for G418 resistance. One of the G418-resistant clones was expanded and termed MSF-T. Both MSF and MSF-T cells were continuously passaged at a 1:2 split ratio, and their growth was expressed as population doubling (PD). (b) The rate of cell division of MSF and MSF-T cells measured by CFSE labeling. MSF cells at 13 PD and MSF-T cells at 79 PD were labeled with CFSE, and their fluorescence intensity was measured at 24, 48, 72 and 120 h post-labeling. A representative of three independent experiments is shown
The growth of MSF cells started to slow down after about 20 PD and ceased altogether around 43 PD. Cells from the MSF-T clone continued to maintain the same doubling time, and this clone was used in subsequent experiments. After 101 doublings, the G418 concentration was reduced to a maintenance dosage of 100 μg/ml, and the cell line was continually propagated over 200 doubling times, where the MSF-T cells continued to grow at the same rate and with no morphological changes. In contrast, large, flat multinucleations started to appear in the untransduced MSF cell cultures (Fig. 2), and the cells stopped dividing after 43 population doublings.
Morphology of MSF, MSF-T and MRC-5 cells in culture. The cells grown to confluency in 6-well plates were photographed at 2X (Panel a) and 20X (Panel b) magnification using a digital camera attached to a phase contrast microscope. MSF cells at 37 PD started to show signs of senescence, such as decreased rate of proliferation and enlarged flattened multinucleated cells. MSF-T cells (37 PD) and MRC-5 cells (12 PD) did not display any morphological changes and showed typical fibroblast shape and contact inhibition. A representative field is shown in each panel
Molecular characterization of life-extended cynomolgus fibroblast cell line MSF-T
To verify that the MSF-T cells express hTERT transcripts, total RNA was extracted from MSF-T cells (at 76 PD) and MSF cells (at 32 PD) and subjected to RT-PCR using hTERT-specific primers. The MSF-T cells were positive for hTERT transcripts, while no hTERT expression was detected in MSF cells (Fig. 3a) and MRC-5 cells (data not shown). As a positive control, hTERT-transduced Telo-RF cells were used. Beta-actin RT-PCR confirmed that comparable amounts of total RNA were used in the experiment.
Telomerase expression in MSF-T cells. (a) hTERT mRNA expression in cynomolgus macaque skin fibroblast cells. Total RNA isolated from MSF (at 12 PD) and MSF-T (at 37 PD) cells were subjected to RT-PCR, and the PCR products were resolved on a 1 % agarose gel. Total RNA isolated from Telo-RF cells was used as the positive control, and primers for β-actin were used to show comparable levels of RNA in each sample. (b) Telomerase activity in MSF (at 12 PD) and MSF-T (at 101 PD) cell extracts were compared using a TRAPeze® Telomerase Detection Kit (Chemicon). The telomerase activity of each cell extract was measured by its ability to extend the TS oligonucleotide provided in the kit. A portion of each cell lysate was also heated at 85 °C for 10 min to inactivate telomerase enzyme activity (HI). The extended oligonucleotides were detected by a PCR generating a ladder of products with 6-nucleotide increments starting at 50 nucleotides. A representative of three independent experiments is shown
To substantiate this evidence, the telomerase activity of MSF and MSF-T cells was assessed using the TRAPeze® Telomerase Detection Assay. The assay measured the ability of telomerase to yield a ladder of products with 6-base pair increments, starting at 50 nucleotides. Cell extracts from MSF-T cells displayed an intense ladder compared to cellular extracts from MSF cells or heat-inactivated MSF-T cells (Fig. 3b).
hTERT-transformed fibroblasts support CyCMV growth
In order to compare the growth of CyCMV in MSF-T, Telo-RF and MRC-5 cells, the cells were infected with CyCMV at two different MOIs (0.3 and 0.03). The cells and supernatants were collected at days 2, 4, 6, 8, 10 and 12 post-infection and viral titers were determined by standard plaque assay using MRC-5 cells (Fig. 4). The virus titer in MSF-T cells infected at 0.3 MOI reached its peak by 6 days post-infection (dpi). The peak titre of the CyCMV-infected MSF-T cells was approximately tenfold lower than the peak viral titre observed in Telo-RF (4 dpi) and MRC-5 (12 dpi) cells, which were comparable to each other (Fig. 4a). The MSF-T cells infected at an MOI of 0.03 reached a maximum viral titer by 8 dpi, and the Telo-RF cells showed peak titers at 6 dpi. CyCMV titers continued to increase beyond 12 dpi in MRC-5 cells infected with CyCMV at an MOI of 0.03. The peak titers in MRC-5 cells infected with CyCMV correlated well with the amount of virus inoculation.
Replication kinetics of CyCMV in MSF-T, Telo-RF and MRC-5 cells. The cells grown in 6-well tissue culture plates were inoculated (in duplicates) with CyCMV at an MOI of 0.3 (a) or 0.03 (b). The cells and the culture supernatant were harvested on days 2, 4, 6, 8, 10 and 12 post-infection, and virus titer in each sample were determined by standard plaque assay on MRC-5 cells. The titers on day “0” in each graph represent the titers of the inoculum. Each data point represents the mean titers from two independent cultures, and the error bars indicate the standard deviation. One of the replicates of MSF-T cells infected at and MOI of 0.03 was spilled during harvest on day 12, and the related data are therefore excluded
CyCMV down-regulates MHC class I in hTERT-transformed fibroblasts
Down-regulation of MHC class I molecules is a salient feature of herpesviruses. We have previously demonstrated that CyCMV down-regulates MHC class I expression in infected MRC-5 cells [3]. The extent of MHC class I down-regulation and the molecular mechanisms involved can be specific to the natural host of a particular herpesvirus. Therefore, we examined MHC class I down-regulation in CyCMV-infected MSF-T cells (Fig. 5). MSF-T, Telo-RF and MRC-5 cells infected with CyCMV for 5 days were harvested, and 100,000 cells from each cell line were subjected to flow cytometric analysis to measure the fully-folded MHC class I molecules on the cell surface using W6/32 monoclonal antibody. MHC class I down-regulation was observed in all three cell lines, and the values for percent down-regulation in the three cell lines (MRC-5 [71.85 % ± 2.65], Telo-RF [52.9 % ± 7.43] and MSF-T [62.75 % ± 7.88]) were comparable (Fig. 5a). The remaining cells were lysed and subjected to immunoblotting using antibodies specific for MHC class I heavy chain (HC) and β2 microglobulin (β2m). When examining the total MHC class I expression, there was a clear decrease in MHC class I HC and β2m levels in all CyCMV-infected cells, and the beta-actin levels were not altered after infection (Fig. 5b).
MHC class I down-regulation in CyCMV-infected MRC-5, Telo-RF and MSF-T cells. (a) The mock- or CyCMV-infected cells were harvested at 5 days post-infection and subjected to flow cytometric analysis following staining with W6/32 mAb. The mock-infected control (dotted line), CyCMV-infected cells (solid line) and the isotype control (gray shaded area) are displayed in a histogram. (b) The same samples were analyzed by western blotting for total MHC class I heavy chain (HC), β2- microglobulin (β2m) and β-actin expression. m, mock-infected; i, CyCMV-infected. A representative of three independent experiments is shown
MSF-T cells are free of Mycoplasma contamination
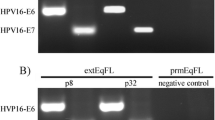
In order to confirm that the established MSF-T cell line was free of mycoplasma contamination, we performed a PCR-based mycoplasma detection assay (Fig. 6). The assay detects all Mycoplasma, Choleplasma and Ureaplasma contaminations to as little as 1-5 fg of their genomic DNA. The internal control template was added to all the samples. The positive control sample gave rise to the expected 267-bp PCR amplicon, while the negative control and the MSF-T supernatant only amplified the internal control (191 bp), confirming that the cell line had no detectable mycoplasma contamination.
Detection of mycoplasma contamination in MSF-T cells by PCR. Cell culture supernatant from MSF-T cells (at 75 PD) was subjected to a highly sensitive PCR-based mycoplasma detection assay. The internal control template was added to all the samples, and positive and negative controls provided in the kit were used. A representative of two independent experiments is shown
Discussion
Cellular replicative senescence occurs as a result of erosions in the telomere DNA [15, 32]. Telomeres protect the natural ends of linear chromosomes, and cellular DNA polymerase is not able to fully duplicate the telomeres during somatic cell division. As a result, 50-200 bp of telomere DNA is lost during each cell division, and these erosions can be prevented by the expression of telomerase, a reverse transcriptase associated with an RNA template that encodes the telomeric repeat sequence [5, 41]. The ectopic expression of human telomerase reverse transcriptase (hTERT) has been used to prevent replicative senescence in a number of different primary cells, including primary fibroblasts [5, 11, 20, 23–25, 30, 35, 42, 43, 45]. Until recently, the most common method of immortalization of primary cells was using viral oncogenes such as simian vacuolating virus 40 (SV40) large T antigen (Tag) or human papillomavirus (HPV) E6 and E7 [7, 18]. These methods result in immortal cell lines; however, the cells exhibit cancer-associated changes such as loss of contact inhibition, differentiation, reduced growth factor requirements, disruption of cell cycle checkpoints and genomic instability. Ectopic expression of hTERT immortalizes many types of primary cells while preserving the characteristics of the parental cells and increases the natural cell repair mechanisms and stress resistance in the transformed cells [21, 27, 29, 31, 38, 42]. While many cell types can be immortalized by ectopic hTERT expression, immortalization of some cell types requires additional changes [13, 22].
Here, we report immortalization of cynomolgus macaque primary skin fibroblast cells by ectopic expression of hTERT. Primary (MSF) cells were transduced with an hTERT expressing VSV-G-pseudotyped Moloney retrovirus, and the transductants were selected based on the G418 resistance. One of the G418-resistant clones was expanded and further characterized. The clone, MSF-T, expressed high levels of hTERT transcripts and telomerase activity. MSF-T cells replicated faster than their untransduced primary counterparts (MSF cells) and showed no morphological changes. MSF-T cells reached over 200 doubling times compared to MSF cells, which reached senescence at 43 PD. In general, a cell line is considered immortalized when it survives three times the life span of its primary counterpart [25]. Therefore, the MSF-T clone can be considered a functionally immortal cell line.
Previous studies have shown that primary human fibroblasts reach senescence between 50 and 72 PD in vitro [14, 19, 22]. MSF cells reached senescence at approximately 43 PD, which was earlier than that of human primary fibroblasts, and the exact reason for the early senescence of MSF cells is not known. According to Kirchoff et al. [23], skin fibroblasts isolated from a rhesus macaque reached senescence at approximately 38 PD, which is consistent with our observations of MSF cells. Therefore, it is possible that primary fibroblasts from non-human primates reach senescence earlier than human primary fibroblasts. The skin fibroblasts reported in our study were isolated from an adult cynomlogus macaque, whereas most of the human primary fibroblasts reported were isolated from young individuals. Therefore, it is possible that the early senescence observed in MSF cells was at least partly due to the age of the donor animal, as telomere lengths are impacted with age. The age of the rhesus macaque used to generate Telo-RF cells was not reported, and thus a direct comparison was not possible [23].
One of the main goals for the development of the MSF-T cell line was to generate a host-specific cell line to propagate the recently isolated cynomolgus-macaque cytomegalovirus (CyCMV) in vitro. Cytomegaloviruses encode a number of cellular tropism genes, which can be spontaneously eliminated from the genome depending on the cell type used for in vitro propagation. The development of a cynomolgus macaque-specific fibroblast cell line will ensure that CyCMV fibroblast tropism genes and host-specific genes are retained in the viral genome.
Human cytomegalovirus is a ubiquitous viral pathogen that causes severe disease in immunocompromised individuals and in newborns. Animal cytomegaloviruses, especially the non-human primate cytomegaloviruses, serve as valuable models for understanding HCMV pathogenesis and for the development of HCMV vaccines and antivirals. Recently, non-human primate cytomegaloviruses emerged as potential vaccine vectors capable of inducing protective immune responses against HIV/SIV [34, 44]. Studies by Louis Picker’s group have shown that RhCMV can be used as a replication-competent viral vector to induce protective immunity against SIV in rhesus macaques [16, 17]. These studies warrant exploration of alternative non-human primate CMV models to confirm their vector potential. The cynomolgus macaque SIV model is one of the most popular alternative HIV vaccine models. With the recently isolation and characterization of cynomolgus macaque cytomegalovirus and the viability of this immortal fibroblast cell line, the cynomolgus macaque SIV model is a promising alternative model for evaluating the potential of CMV-based SIV/HIV vaccines.
Previous studies have shown that there are no appreciable differences in CMV viral growth kinetics when comparing primary (untransduced) and transduced fibroblast cell lines [6, 8, 10, 28]. Additional studies have confirmed that the presence of hTERT in immortalized cells does not affect CMV gene expression [1, 8, 28]. Human studies have demonstrated that the hTERT-transduced MRC-5 cells are fully permissive to HCMV, and peak virus titers are comparable in both cell types. However, the rate of HCMV proliferation is slightly slower in hTERT-immortalized MRC-5 cells compared to the primary MRC-5 cells [28]. In contrast, RhCMV replicates slightly faster in hTERT transduced Telo-RF cells than in the primary rhesus fibroblasts [8]. This difference was more apparent when the cells were infected at a low MOI. Although we did not directly compare MSF-T cells to the primary isolated cells, primary MSF cells are fully permissive to CyCMV infection and exhibit characteristic CMV plaque formation (data not shown). Alternatively, in the experiments described herein, we have compared the immortalized MSF-T cell line to other cell lines (Telo-RF and MRC-5) that are routinely used for the propagation of CMV and for CMV-related studies. Our experiments clearly show that MSF-T cells support the growth of CyCMV; however, the peak viral titers and rates of viral replication differ in the three different cell lines used. CyCMV replicated faster in Telo-RF cells than in MSF-T and MRC-5 cells. However, the peak viral titer of CyCMV in MRC-5 cells was comparable to that in Telo-RF cells when the cells were inoculated at a higher MOI. The replication rate of CyCMV was slowest in the MRC-5 cells. A similar replication pattern was also observed when the cells were infected at a lower MOI. The exact reason for the observed differences in viral replication and peak viral titers is not clear. However, based on observations by Chang et al. [8], we believe that the rate of virus replication is directly related to the rate of cell proliferation. Data from our CFSE labelling experiment showed that MSF-T cells proliferate faster than MSF cells and that MSF cells have a doubling time similar to MRC-5 cells. Subsequent CFSE-labelling experiments showed that Telo-RF cells proliferate slightly faster than MSF-T cells (data not shown). Therefore, we believe that the intermediate proliferation rate of CyCMV observed in MSF-T cells is directly related to the intermediate replication rate of MSF-T cells.
The peak viral titers differed between the three fibroblast cell lines. Telo-RF cells infected with CyCMV at MOI of 3 reached maximum viral titer by 4 days post-infection and began gradually decreasing thereafter. Directly related to this pattern, Telo-RF cells replicated faster and reached confluency by 3-4 days post-inoculation and subsequently showed a reduced replication rate due to contact inhibition. MRC-5 cells replicated slower than Telo-RF and MSF-T cells and did not show any signs of overcrowding even at 10-12 days post-inoculation. We believe that this allowed CyCMV to replicate at a slower rate and to continue beyond 10 days so that the peak virus titers in MRC-5 cells were comparable to the peak viral titers achieved in the Telo-RF cells. MSF-T cells replicate at a slower rate than Telo-RF cells and at a faster rate than MRC-5 cells, thus displaying intermediate viral growth kinetics. When the cells were infected with tenfold less virus (MOI 0.3), the differences in viral titers and replication rate became more obvious. Taken together, we believe that the observed differences in the CyCMV replication rate and the peak viral titre were directly related to the differences in the rate of cell proliferation in the three cell types examined.
The reason for the observed difference in the rate of cell proliferation between hTERT-transduced MSF-T and Telo-RF cells was not clear. Both cell types were transduced with the same hTERT construct; however, the site of integration of the hTERT gene into the cellular genome may be different in the two cell types and may therefore alter the phenotype of the transduced cell lines. Mycoplasma contamination of cell cultures is a widespread problem, and one of the earliest signs of mycoplasma contamination is reduced rate of cell proliferation. We have confirmed that MSF-T cells are free of mycoplasma contamination by PCR, one of the most sensitive mycoplasma detection methods available. We have also examined MSF-T cells by electron microscopy, and no mycoplasma contamination was observed (Ambagala et al., unpublished data).
Previously, we have shown that CyCMV down-regulates MHC class I expression in MRC-5 cells. Based on the genomic sequence, CyCMV encodes at least four viral proteins, cyUS2, cyUS3, cyUS11 and cy203, that are able to interfere with the MHC class I antigen presentation pathway [26]. Viral immunevasins can be species-specific and therefore can have a different effect on cells from different species. We compared the MHC class I down-regulation in CyCMV-infected MSF-T cells to that in CyCMV-infected MRC-5 and Telo-RF cells. In addition to the MHC class I expression on the cell surface, we also compared the total MHC class I heavy chain (HC) and β2 microglobulin (β2m) expression in CyCMV-infected and mock-infected cells. MHC class I down-regulation was evident in all three cells types, and the extent of down-regulation on the cell surface and the HC and β2m levels were comparable in all three cell types. Therefore, CyCMV MHC class I evasion genes appear to function similarly in human, rhesus, and cynomolgus fibroblasts in vitro.
Telo-RF cells are readily transfected with lipid-based transfection agents (TransIT from Mirus Bio LLC, Madison, WI and Fugene 6 from Roche) compared to their primary counterparts [8]. We transfected MSF-T cells with a plasmid expressing enhanced green fluorescent protein (pEGFP-N1, Clontech laboratories) using a similar approach (Lipofectamine-2000, Invitrogen Inc.). In comparison to MSF cells, we did not observe an increase in transfection efficiency in the MSF-T cells (data not shown).
In summary, we have generated an immortalized cynomolgus macaque dermal fibroblast cell line by ectopic expression of hTERT. The cell line, MSF-T, is fully permissive to CyCMV infection, has reached over 200 PD, and continues to divide without any morphological changes. This cell line will be beneficial for in vitro research related to cynomolgus macaques and for propagating CyCMV in a host-specific cell line. Furthermore, MSF-T cells may be used to propagate other herpesviruses, such as varicella zoster virus or rhadinoviruses, that may be isolated from cynomolgus macaques in the future.
References
Adair R, Liebisch GW, Lerman BJ, Colberg-Poley AM (2006) Human cytomegalovirus temporally regulated gene expression in differentiated, immortalized retinal pigment epithelial cells. J Clin Virol 35:478–484
Akari H, Mori K, Terao K, Otani I, Fukasawa M, Mukai R, Yoshikawa Y (1996) In vitro immortalization of Old World monkey T lymphocytes with Herpesvirus saimiri: its susceptibility to infection with simian immunodeficiency viruses. Virology 218:382–388
Ambagala AP, Marsh A, Chan J, Pilon R, Fournier J, Mazzulli T, Sandstrom P, Willer DO, MacDonald KS (2011) Isolation and characterization of cynomolgus macaque (Macaca fascicularis) cytomegalovirus (CyCMV). Virology 412:125–135
Baroncelli S, Negri DR, Michelini Z, Cara A (2008) Macaca mulatta, fascicularis and nemestrina in AIDS vaccine development. Expert Rev Vaccines 7:1419–1434
Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE (1998) Extension of life-span by introduction of telomerase into normal human cells. Science 279:349–352
Bresnahan WA, Hultman GE, Shenk T (2000) Replication of wild-type and mutant human cytomegalovirus in life-extended human diploid fibroblasts. J Virol 74:10816–10818
Bryan TM, Reddel RR (1994) SV40-induced immortalization of human cells. Crit Rev Oncog 5:331–357
Chang WL, Kirchoff V, Pari GS, Barry PA (2002) Replication of rhesus cytomegalovirus in life-expanded rhesus fibroblasts expressing human telomerase. J Virol Methods 104:135–146
Chen WH, Wang XX, Lin W, He XW, Wu ZQ, Lin Y, Hu SN, Wang XN (2006) Analysis of 10,000 ESTs from lymphocytes of the cynomolgus monkey to improve our understanding of its immune system. BMC Genomics 7:82
Compton T (1993) An immortalized human fibroblast cell line is permissive for human cytomegalovirus infection. J Virol 67:3644–3648
Condon J, Yin S, Mayhew B, Word RA, Wright WE, Shay JW, Rainey WE (2002) Telomerase immortalization of human myometrial cells. Biol Reprod 67:506–514
Condreay JP, Condreay LD, Huber BE (1997) Effects of epidermal growth factor on growth of cell lines established from cynomolgus monkey hepatocytes with SV40 T antigen. In Vitro Cell Dev Biol Anim 33:228–231
Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA, Louis DN, Li FP, Rheinwald JG (2000) Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol 20:1436–1447
Franco S, MacKenzie KL, Dias S, Alvarez S, Rafii S, Moore MA (2001) Clonal variation in phenotype and life span of human embryonic fibroblasts (MRC-5) transduced with the catalytic component of telomerase (hTERT). Exp Cell Res 268:14–25
Greider CW (1990) Telomeres, telomerase and senescence. Bioessays 12:363–369
Hansen SG, Vieville C, Whizin N, Coyne-Johnson L, Siess DC, Drummond DD, Legasse AW, Axthelm MK, Oswald K, Trubey CM, Piatak M Jr, Lifson JD, Nelson JA, Jarvis MA, Picker LJ (2009) Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med 15:293–299
Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, Whizin N, Oswald K, Shoemaker R, Swanson T, Legasse AW, Chiuchiolo MJ, Parks CL, Axthelm MK, Nelson JA, Jarvis MA, Piatak M Jr, Lifson JD, Picker LJ (2011) Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473:523–527
Hawley-Nelson P, Vousden KH, Hubbert NL, Lowy DR, Schiller JT (1989) HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. Embo J 8:3905–3910
Hayflick L, Moorhead PS (1961) The serial cultivation of human diploid cell strains. Exp Cell Res 25:585–621
He YL, Wu YH, He XN, Liu FJ, He XY, Zhang Y (2009) An immortalized goat mammary epithelial cell line induced with human telomerase reverse transcriptase (hTERT) gene transfer. Theriogenology 71:1417–1424
Jiang XR, Jimenez G, Chang E, Frolkis M, Kusler B, Sage M, Beeche M, Bodnar AG, Wahl GM, Tlsty TD, Chiu CP (1999) Telomerase expression in human somatic cells does not induce changes associated with a transformed phenotype. Nat Genet 21:111–114
Kim HS, Shin JY, Yun JY, Ahn DK, Le JY (2001) Immortalization of human embryonic fibroblasts by overexpression of c-myc and simian virus 40 large T antigen. Exp Mol Med 33:293–298
Kirchoff V, Wong S, St JS, Pari GS (2002) Generation of a life-expanded rhesus monkey fibroblast cell line for the growth of rhesus rhadinovirus (RRV). Arch Virol 147:321–333
Kitagawa M, Ueda H, Iizuka S, Sakamoto K, Oka H, Kudo Y, Ogawa I, Miyauchi M, Tahara H, Takata T (2007) Immortalization and characterization of human dental pulp cells with odontoblastic differentiation. Arch Oral Biol 52:727–731
Lee KM, Nguyen C, Ulrich AB, Pour PM, Ouellette MM (2003) Immortalization with telomerase of the Nestin-positive cells of the human pancreas. Biochem Biophys Res Commun 301:1038–1044
Marsh AK, Willer DO, Ambagala AP, Dzamba M, Chan JK, Pilon R, Fournier J, Sandstrom P, Brudno M, MacDonald KS (2011) Genomic sequencing and characterization of cynomolgus macaque cytomegalovirus. J Virol 85:12995–13009
Mazzucchelli GD, Gabelica V, Smargiasso N, Fleron M, Ashimwe W, Rosu F, De Pauw-Gillet MC, Riou JF, De Pauw E (2008) Proteome alteration induced by hTERT transfection of human fibroblast cells. Proteome Sci 6:12
McSharry BP, Jones CJ, Skinner JW, Kipling D, Wilkinson GW (2001) Human telomerase reverse transcriptase-immortalized MRC-5 and HCA2 human fibroblasts are fully permissive for human cytomegalovirus. J Gen Virol 82:855–863
Morales CP, Holt SE, Ouellette M, Kaur KJ, Yan Y, Wilson KS, White MA, Wright WE, Shay JW (1999) Absence of cancer-associated changes in human fibroblasts immortalized with telomerase. Nat Genet 21:115–118
Morales CP, Gandia KG, Ramirez RD, Wright WE, Shay JW, Spechler SJ (2003) Characterisation of telomerase immortalised normal human oesophageal squamous cells. Gut 52:327–333
Nakamura H (2008) hTERT-immortalized cells useful for analyzing effects of low-dose-rate radiation on human cells. J Radiat Res 49:9–15
Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR (1997) Telomerase catalytic subunit homologs from fission yeast and human. Science 277:955–959
O’Connor DH (2006) Chinese rhesus and cynomolgus macaques in HIV vaccine and pathogenesis research. Future Virology 1:165–172
Picker LJ, Hansen SG, Lifson JD (2012) New paradigms for HIV/AIDS vaccine development. Annu Rev Med 63:95–111
Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, Peyton M, Zou Y, Kurie JM, Dimaio JM, Milchgrub S, Smith AL, Souza RF, Gilbey L, Zhang X, Gandia K, Vaughan MB, Wright WE, Gazdar AF, Shay JW, Minna JD (2004) Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res 64:9027–9034
Stam NJ, Spits H, Ploegh HL (1986) Monoclonal antibodies raised against denatured HLA-B locus heavy chains permit biochemical characterization of certain HLA-C locus products. J Immunol 137:2299–2306
Suemori H, Tada T, Torii R, Hosoi Y, Kobayashi K, Imahie H, Kondo Y, Iritani A, Nakatsuji N (2001) Establishment of embryonic stem cell lines from cynomolgus monkey blastocysts produced by IVF or ICSI. Dev Dyn 222:273–279
Takano H, Murasawa S, Asahara T (2008) Functional and gene expression analysis of hTERT overexpressed endothelial cells. Biologics 2:547–554
Takaoka T, Katsuta H, Endo M, Sato K, Okumura H (1962) Establishment of a cell strain, JTC-12, from cynomolgus monkey kidney tissue. Jpn J Exp Med 32:351–368
Titti F, Zamarchi R, Maggiorella MT, Sernicola L, Geraci A, Negri DR, Borsetti A, Menin C, D’Andrea E, Modesti A, Masuelli L, Verani P, Chieco-Bianchi L, Amadori A (2002) Infection of simian B lymphoblastoid cells with simian immunodeficiency virus is associated with upregulation of CD23 and CD40 cell surface markers. J Med Virol 68:129–140
Wyllie FS, Jones CJ, Skinner JW, Haughton MF, Wallis C, Wynford-Thomas D, Faragher RG, Kipling D (2000) Telomerase prevents the accelerated cell ageing of Werner syndrome fibroblasts. Nat Genet 24:16–17
Yang J, Chang E, Cherry AM, Bangs CD, Oei Y, Bodnar A, Bronstein A, Chiu CP, Herron GS (1999) Human endothelial cell life extension by telomerase expression. J Biol Chem 274:26141–26148
Yudoh K, Matsuno H, Nakazawa F, Katayama R, Kimura T (2001) Reconstituting telomerase activity using the telomerase catalytic subunit prevents the telomere shorting and replicative senescence in human osteoblasts. J Bone Miner Res 16:1453–1464
Yue Y, Barry PA (2008) Rhesus cytomegalovirus a nonhuman primate model for the study of human cytomegalovirus. Adv Virus Res 72:207–226
Zou Y, Yi X, Wright W, Shay J (2002) Human telomerase can immortalize Indian muntjac cells. Exp Cell Res 281:63–76
Acknowledgments
We would like to thank the veterinary and technical staff at the animal NHP colony of Health Canada, Ottawa, and Drs. Tania Watts and Laurent Sabbagh for the pME-VSV-G and Phoenix cells. A.A. was supported by a postdoctoral fellowship from the Ontario HIV Treatment Network (OHTN), and A.K.M. was supported by the Queen Elizabeth II/Community Health Graduate Scholarships in Science and Technology and the Bernhard Cinader Graduate Scholarship in Immunology. D.O.W was supported by a Junior Investigator Development Award from the OHTN, and KSM holds a senior scientist award from the OHTN. This research was partly funded by Canadian Institutes of Health Research (CHIR) and the Canadian Foundation for AIDS research (CANFAR).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ambagala, A.P., Marsh, A.K., Chan, J.K. et al. Establishment of an immortal cynomolgus macaque fibroblast cell line for propagation of cynomolgus macaque cytomegalovirus (CyCMV). Arch Virol 158, 955–965 (2013). https://doi.org/10.1007/s00705-012-1568-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-012-1568-4