Abstract
Oceanic islands have isolated biota, which typically include many endemic species. However, island endemics are vulnerable due to small population sizes, and they are often threatened by habitat destruction or by introduced pests and predators. Adequate conservation planning requires good information on genetic variability and population structure, also when seemingly viable species are considered. Here, I analysed the genetic structure in the terrestrial orchid Dactylorhiza foliosa, which is endemic to Madeira. This species is a characteristic component of evergreen laurel forests occupying the northern slopes of the island. Levels of diversity in both the plastid genome and in the nuclear genome were comparable to levels of diversity found in congeners growing in continental regions. Within populations, plants separated by distances up to 256 m shared plastid haplotypes significantly more often than plants at random, but when nuclear markers were considered, only plants growing closer than eight metres were significantly more closely related. Analysis of plastid marker variation revealed that gene dispersal by seeds is not sufficiently large to counterbalance the accumulation of mutations that build up divergence between the most distant populations. However, differentiation in the nuclear genome was considerably smaller, suggesting that gene dispersal by pollen is much more efficient than gene dispersal by seeds in D. foliosa. The overall pollen to seed dispersal ratio, mp/ms, was 7.30. Considering genetic parameters, conditions for long-term persistence of D. foliosa on Madeira seem to be good.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants and animals inhabiting isolated islands have long fascinated travellers and students in natural history. Islands hold disproportionally high numbers of endemic species compared to continental regions, and the numbers of endemics per unit surface area may be extremely high (Myers et al. 2000; Kiera et al. 2009; Veron et al. 2019). Studies of island biota inspired Wallace and Darwin to formulate the theory of evolution by natural selection (Cox et al. 1976), and islands provide excellent opportunities to study various aspects of speciation.
Species numbers on oceanic islands can be elevated as result of niche differentiation. Factors promoting such differentiation include climate variability associated with altitudinal gradients, expressed as e.g. differences in precipitation, temperature or predomint wind directions. A high species diversity can also depend on high speciation rates, which can be elevated as an effect of small population sizes (Cox et al. 1976; Irl et al. 2017). Organismal groups on archipelagos of oceanic islands may additionally have expanded the number of species by the process of adaptive radiation, i e., by diversification into multiple niches when colonizing new islands, often multiplied by back-colonization of islands where the lineage is already represented by ancestral forms.
Notwithstanding their importance, island biota occupy small areas and must be considered as vulnerable in a conservation context. Plants and animals inhabiting isolated islands typically (i) comprise restricted numbers of individuals, and (ii) contain reduced levels of genetic diversity as compared to related species living in mainland regions (Hamrick and Godt 1990; Frankham 1997). A small population size leads to an increased risk of extinction due to habitat destruction or stochastic fluctuations in numbers of individuals (Lande 1999), whereas a restricted genetic diversity may impede adaptive responses to environmental change in the future (Frankel and Soulé 1981; Frankham 1997). Many island species are narrow endemics and their island habitats have often been subject to habitat destruction to a much higher degree than corresponding mainland habitats. Furthermore, island species are often threatened by predators, pests or competitors unintentionally or intentionally introduced by man. For all these reasons, many island species are threatened and of concern in conservation (Frankham 1997).
Whereas data on population sizes can be sensored by ordinary methods of observation, data on genetic diversity are difficult to obtain and are lacking for most island organisms. However, genetic data are necessary for estimating levels of population subdivision, local migration patterns, gene flow and other parameters useful for conservation planning. Genetic variation is a prerequisite for biological organisms to respond to future changes in the environment, including effects of climate warming. For island organisms, genetic diversity is especially important, since these organisms are stuck where they are and do not have the option to migrate along climatic gradients as the global climate is successively changing.
In the present study, I analysed the genetic structure in the orchid Dactylorhiza foliosa (Soland. ex Lowe) Soó, which is restricted to the island of Madeira, Portugal. The island is situated in the Atlantic Ocean about 845 km SW of the Iberian Peninsula. On the island, D. foliosa is widespread in the evergreen laurel forest, “Laurisilva” covering the northern slopes at altitudes between ca 500 and 1100 m a. s. l. (Delforge 2001; Baumann et al. 2006; Capelo et al. 2005). Here, it grows most frequently in semi-open habitats, and is common on rocky slopes, on roadsides, along paths, and along the water-channels, “levadas”, which run horizontally along the mountain slopes and which lead water to cultivated areas on the drier south-facing slopes of the island. The orchid also has scattered occurrences in the slightly drier habitats above and below the laurel forest zone at altitudes between ca. 300 to 1500 m a. s. l. (Jardim and Francisco 2000; Delforge 2001; Baumann et al. 2006; Rankou 2011).
The plant is a herbaceous perennial between 20 and 80 cm tall. The leaves are more or less linear, shining, usually unspotted and aggregated along the lower portion of the stem. The inflorescence contains about 10–50 pinkish flowers with a lip of ca 8–16 × 9–18 mm covered with spots and short streaks (Jardim and Francisco 2000; Delforge 2001; Baumann et al. 2006). The plant usually forms discrete individuals with one or a few flowering stems from an underground tuber, but sometimes whole clumps of plants are formed as the result of successive division of the tuber.
Dactylorhiza foliosa is a diploid member of the D. maculata s.l. species complex (Delforge 2001). Molecular studies show that the closest relative to D. foliosa is the western European subpopulation of D. maculata subsp. maculata (Hedrén et al. 2001; Bateman et al. 2003; Devos et al. 2005; Brandrud et al. 2020). However, D. maculata subsp. maculata is invariably tetraploid, and the distinct morphology of D. foliosa suggests that it has been separated from the mainland populations of D. maculata for a long period of time. Other members of the D. maculata complex are known from the mountain regions of NW Africa (Delforge 2001), but these taxa are poorly studied genetically. The complex is lacking from other Macaronesian islands.
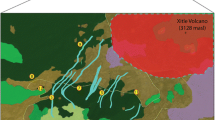
Available data on D. foliosa are inconclusive for estimating its evolutionary potential and adaptability. On the one hand, the orchid is restricted to the ca 150 km2 of laurel forests on Madeira, and it is not known from anywhere else, which suggests that the genetic diversity in the species might be restricted. On the other hand, the orchid is widespread within the laurel forest habitat, including secondary vegetation (Baumann et al. 2006), and the population size is large (Rankou 2011), which suggests that the species could store a considerable amount of genetic diversity. Moreover, laurel forests covers much of the northern slopes of Madeira without any major gaps and the geographic subpopulations are probably well connected (Fig. 1), which should allow for genetic connectivity and good opportunities for the maintenance of the genetic diversity that exists within the species.
Map showing sampling sites for Dactylorhiza foliosa and the distribution of evergreen Laurel forest (shaded) on Madeira. Distribution data reproduced from Secretaria Regional do Ambiente e Recursos Naturais, https://ifcn.madeira.gov.pt/18-sem-categoria/163-laurissilva-da-madeira-2.html; accessed 26 Feb 2019
Similar to other species of Dactylorhiza, D. foliosa is characterised by efficient mechanisms for seed and pollen dispersal. Like most orchids, D. foliosa has minute, partly air-filled seeds that can be transported long distances by wind. The flowers contain no reward but act by deceit. The syrphid fly Eristalix tenax and the bumble-bee Bombus maderensis have been observed as the most frequent pollinators (Fernandes et al. 2005).
A better understanding of genetic diversity in island endemics is interesting in itself and may also be important for implementation of conservation strategies. In the present study, I have chosen to analyse variation in both plastid markers and nuclear markers in D. foliosa. By combining data from the resulting data sets, it is possible to compare the effects of gene dispersal by seeds and gene dispersal by pollen. By sampling populations from different localities on Madeira and by mapping physical distance between individuals within sampling localities, gene dispersal and genetic diversity structure could be described for both the species level and the population level. Finally, I compare the results obtained for D. foliosa with similar data obtained from previously analysed species of Dactylorhiza, and ask whether the genetic structure in D. foliosa would be of concern in a conservation context.
Materials and methods
Material and sampling
Samples of D. foliosa were taken from eight sites, separated by between 0.7 and 26 km (Online resource 1). Plants were sampled along levadas, paths or roadsides. Three to forty plants were sampled from each site, depending on the density and extent of the local population. To avoid repeated sampling of the same genet, samples were taken from plants separated by at least one meter. The average distance between adjacent samples within sites ranged between 12.8 and 151.2 m (Online resource 1). The minimum distance between samples within sites were 1 or 2 m, whereas the maximum distance ranged between 71 and 1051 m. Some populations were much larger than the sampling areas and might be connected without any major gaps in distribution to adjacent populations, for instance the populations at Portela and Ribeiro Frio.
A few flowers or a small amount of leaf tissue was collected for DNA extraction, leaving the rest of the plants for continued growth. The sampled material was dried in silica gel (Chase and Hills 1991) and stored at room temperature until extraction. DNA was extracted from one dried flower with bract or from ca 1 cm2 dried leaf tissue according to the 2 × CTAB procedure (Doyle and Doyle 1990). From one individual per population an additional flower was sampled and deposited in the Lund botanical museum (LD) as voucher material.
Molecular analyses
To enable discrimination between plastid genomes, ten repeat regions described in Hedrén et al. (2008) were initially screened for variation; 1, 4, 5, 6, 6B, 8, 9, 10b, 11b, 17, 18, and 19B (Online resource 2). After initial screening, the four regions 6B, 10b, 11b, and 19B were selected for analysis of the entire material. Locus 6B is a complex indel region, 10b is a polyA-polyTA-polyT repeat region and 11b and 19B are mononucleotide repeat regions (Online resource 2). Each combination of fragment sizes at the four repeat regions was recognized as a separate haplotype. Finally, a haplotype distance matrix for downstream analyses was constructed by counting the number of repeat regions that differed in fragment size between any two haplotypes.
The ITS region was studied as in Pillon et al. (2007), in which ITS alleles were identified by their combined fragment lengths at two size-variable regions (Online resource 2). ITS has previously been shown to vary in the D. maculata s.l. complex, in some cases even within taxa (Devos et al. 2005; Pillon et al. 2007; Ståhlberg and Hedrén 2010).
Variation in the nuclear genome was studied by analysis of four microsatellite loci (Online resource 2). Two of these, ms3 and ms11 gave rise to fragments separated by steps of three base-pairs, whereas ms2 and D2501 gave rise to fragments separated by steps of two base-pairs. The first two loci were described by Nordström and Hedrén (2007), and the latter two were added in Hedrén et al. (2018).
All PCR reactions were performed in a reaction volume of 6.4 μL containing 4.4 μL deionized and sterilized H2O, 0.66 μL 10 × reaction buffer (100 mM Tris–HCl pH 8.3, 500 mM KCl, 15 mM MgCl2), 0.56 μL dNTPs (2.5 mM of each nucleotide), 0.25 μL Cy5-labelled primer (10 pg/μL), 0.1 μL unlabelled complementary primer (25 pmol/μL), 0.03 μL AmpliTaq polymerase (5 u/μL; Applied Biosystems), and 0.6 μL template DNA (14 ng/μL).
Size variable fragments in the plastid genome were amplified by an initial round of denaturing at 94 °C for 2 min, followed by 40 cycles of 94 °C for 1 min, 50–58 °C (Online resource 2) for 1 min, 72 °C for 1 min 30 s, ended by a final extension of 72 °C for10 min. Nuclear microsatellite loci were amplified by an initial denaturing at 94 °C for 3 min, followed by 36 cycles of 94 °C for 30 s, annealing at a temperature 54 °C for 30 s, extension at 72 °C for 45 s, and concluded by a final extension period at 72 °C for 10 min. The ITS alleles were amplified by means of the same program, but with an annealing temperature of 52 °C.
The PCR product from each reaction was mixed with 8.5 μl formamide containing appropriate size markers to enable exact relative size determination of the amplified fragments. The fragments were then heated at 94 °C for ca 20 min before loading on a horizontal acrylamide gel and separated by size on an ALFexpress II automated sequencer (Amersham Pharmacia Biotech). Fragment sizes were determined by using ALFwin™ fragment analyser 1.03.01 software (Amersham Pharmacia Biotech).
Data analysis
For each population sample, the following statistics were calculated on basis of nuclear microsatellite data (Table 1): A, average number of alleles per locus; Ae, effective number of alleles per locus; AR, allelic richness adjusted for a population size of 5 samples (10 gene copies); He, expected diversity corrected for sample size; and FIS, inbreeding coefficients calculated according to Weir and Cockerham (1984).
For plastid haplotype data, the following parameters were calculated for each population sample: A, number of different haplotypes; Ae, effective number of haplotypes; AR, haplotype richness adjusted for a population size of 5 samples (5 haplotype copies); and He, expected diversity corrected for sample size (Table 1). All statistics were obtained in SPAGeDi 1.4 (Hardy and Vekemans 2002).
The general differentiation between population samples was described by FSTs calculated according to an ANOVA approach as described Weir and Cockerham (1984). Separate analyses were performed for the plastid and nuclear data sets. To test for an isolation by distance structure (Wright 1943), the differentiation between pairs of populations were calculated and compared to geographic distances by means of Mantel tests (Table 2). Differentiation was given as pairwise FST/(1—FST) and geographic distances as the logarithm of geographic distance (Rousset 1997). I also tested for an association between pairwise FST derived from nuclear data with pairwise GST from the haplotype data. Pairwise genetic distances were calculated in SPAGeDi 1.4 (Hardy and Vekemans 2002). Associations between distance matrices were tested by10000 rounds of permutation in NTSYS-pc 2.2 (Rohlf 2005).
To analyse D. foliosa for patterns of genetic differentiation, I constructed a maximum parsimony network for the plastid haplotypes using the computer program Network 10.2.0.0 (Fluxus technology 2007), and I investigated the microsatellite data for potential clustering of nuclear genotypes using the computer program Structure 2.3.4 (Pritchard et al. 2000). For the plastid loci, I assumed a stepwise mutation model and weighted all mutations equally, irrespective of the amount of variability shown at the different marker sites. For the nuclear data, I used the mixed ancestry model, assumed independent allele frequencies, and made no a priori assumptions of population belonging. I tested the data for separation into K = 1 to 10 clusters and repeated the analysis 20 times for each value of K. I used a burnin period of 10,000 steps and made 10,000 iterations in each run. The optimum number of clusters was estimated by the Evanno method (Evanno et al. 2005) as implemented in the computer program Structure Harvester (Earl and vonHoldt 2012), and the optimal combined solution for the multiple runs of each value for K was estimated by the computer program CLUMPP (Jakobsson and Rosenberg 2007).
To test for a phylogeographic structure, i.e. whether different haplotypes or microsatellite alleles are significantly more divergent between populations than within populations (Hardy and Vekemans 2002), I compared differentiation measures that describe the degree of differentiation between markers with values obtained by permutation of the original data. For the plastid haplotype data the differentiation measure NST was calculated (Pons and Petit 1996). Here, genetic divergence was estimated as the number of loci with different fragment sizes between any pair of haplotypes. A corresponding GST, along with 95% confidence intervals, was obtained by 10,000 rounds of permutation of genetic distances in the haplotype distance matrix. For the nuclear microsatellites. I estimated the statistic RST, which is based on the numbers of mutational steps between microsatellite alleles (Slatkin 1995; Michalakis and Excoffier 1996). A corresponding FST, along with 95% confidence intervals, was estimated by 10,000 rounds of permutation of allele sizes among alleles within loci (see also Hardy and Vekemans 2002). Calculations were performed in SPAGeDi 1.4 (Hardy and Vekemans 2002).
The FST and the FIS estimates from nuclear microsatellites and the FST from plastid haplotypes were used to calculate the pollen to seed-flow ratio, mp/ms according to the formula given by Ennos (1994) and Petit et al. (2005):
where FST(B) and FIS(B) are derived from biparentally inherited microsatellite markers and FST(M) is derived from maternal plastid markers.
The spatial genetic structure, SGS, within populations was estimated by use of the kinship coefficient Fij of Loiselle et al. (1995). I calculated mean kinship between individuals within distance intervals of 4, 8, 16, 32, 64, 128 and 256 m, which subdivided the material into categories containing appropriate numbers of pairwise comparisons, and analysed whether kinship decreased with distance. To test whether the kinship values were significant, I compared the observed values with the mean values obtained by 10,000 rounds of permutation of individual locations among all individuals. These results were also used to calculate the Sp statistic of Vekemans and Hardy (2004):
bF is the linear regression slope of Fij (n) on the natural logarithm of the distance intervals, and Fij (1) is the mean kinship coefficient for the first distance interval.
Separate analyses were performed for plastid haplotype and nuclear microsatellite data. All within-population statistics were calculated in SPAGeDi 1.4 (Hardy and Vekemans 2002).
Results
A total of 33 plastid haplotypes was found in D. foliosa (Online resource 3). The small population of Encumeada West-Central comprising only three samples, was found with a single haplotype (Table 1, Online resource 4). In the remaining populations, between four (Encumeada West) and 18 (Encumeada North) haplotypes were encountered within populations. Correcting for sample size by rarefaction with k = 10 (excluding Encumeada West-Central and Rabaçal), between 3.82 (Encumeada West) and 7.53 (Encumeada North) haplotypes per populations was estimated (Table 1). Expected haplotype diversity (He) ranged from 0.0 (Encumeada West) and 0.69 (Ribeiro Frio) to 0.93 (Encumeada N).
In ITS all plants were fixed for the ITS allele designated as ITS-I in Pillon et al. (2007).
A total of 29 different alleles were encountered at the four examined nuclear microsatellite loci (Online resource 5). Locus ms13 was fixed for an allele of 90 bp in all populations. At the remaining loci, three alleles were found at ms 11 with between one and two alleles per population, six alleles at ms 14 with between two and four alleles per population, and 19 alleles at locus D2501 with between three and fifteen alleles per population (Online resource 5). Adjusting for sample size by rarefaction and comparing all populations with the population with the lowest number of individuals (Encumeada VC; n = 3, six gene copies), the number of alleles per population ranged from 1.00 to 1.53 at ms11, 1.25–2.97 at ms14, and 3.00 to 5.10 at D2501 (Online resource 5). The expected gene diversity (He) ranged from 0.35 (Encumeada West) and 0.391 (Pico das Pedras) to 0.575 (Rabaçal) (Table 1, Online resource 5).
The global inbreeding coefficient over populations and loci was moderate in D. foliosa, with FIS = 0.092 (Table 3). Inbreeding coefficients varied over populations, ranging from values close to zero in the population samples from Ribeiro Frio and Portela to FIS = 0.8 and FIS = 0.523, respectively, in the small samples from Encumeada West-Central and Rabaçal (Table 1). The average unweighted FIS over populations samples was 0.222 (Table 1). Disregarding the smallest population samples, the average inbreeding coefficient in the remaining six samples was 0.075.
The overall FST for plastid haplotypes was 0.137 (Table 3). A clearly lower amount of differentiation between populations was observed in the nuclear genome and the overall FST estimated on basis of nuclear microsatellites was 0.018.
Comparison of pairwise FST values derived from plastid haplotype data and geographic distances between sampling sites by Mantel tests (Table 2) did not reveal presence of any significant isolation by distance structure, IBD (Z = 0.716, p (random Z ≥ obs Z) = 0.204). For the nuclear genome, comparison of pairwise FST values derived from microsatellite allele frequency data and geographic distances between sampling sites did also not reveal any significant isolation by distance structure (Z = -0.173, p (random Z ≥ obs Z) = 0.758). A somewhat stronger correlation was found between pairwise FSTs derived from plastid haplotypes and pairwise FSTs derived from nuclear microsatellites, but the association was still not significant (Z = 0.952, p (random Z ≥ obs Z) = 0.102).
The relationships of the encountered plastid haplotypes were investigated in a parsimony network analysis. Eight most parsimonious trees of 40 steps each were found. The resulting network is given as Fig. 2. To some extent, the more common haplotypes were often shared between neighbouring sites, or else neighbouring sites often shared related haplotypes with each other.
Parsimony network of the 33 plastid haplotypes encountered in Dactylorhiza foliosa. The network is based on eight individual unrooted trees of 40 steps each. Haplotypes are represented by pie diagrams in which geographic origin is given by different colours. The sizes of the pie diagrams are proportional to the numbers of samples carrying a particular haplotype. The directions of the edges linking haplotypes correspond to mutations in different markers. The unit length of the edge is one mutational step unless otherwise indicated. Haplotypes are labelled as in online resources 3 and 4. Small filled diamonds denote median vectors
To test D. foliosa for presence of any substructure on basis of nuclear genomes, nuclear microsatellite genotypes were analysed for aggregation into one to ten (K) groups by means of Bayesian cluster analysis as implemented in the computer program Structure. However, whatever the number of K, each sample invariably obtained about equal probability of belonging to any of the groups tested for (Online resources 6). Applying the Evanno method to obtain the optimal number of subgroups, K = 4 had a slightly higher probability than K = 2 (Online resources 7), but none of these solutions were visibly any better than any of the other solutions.
Calculation of NST and comparison with the corresponding GST obtained by permutation of numbers of differences between pairs of haplotypes in the haplotype distance matrix (Table 3), revealed the presence of a significant general phylogeographic structure in the plastid genome of D. foliosa (0.400 vs 0.229; p < 0.000***).
In contrast, comparison of the RST, which considers the number of mutational steps between microsatellite alleles at locus, and the corresponding FST, obtained by permutation of allele sizes among alleles within each locus, revealed no significant phylogeographic structure in the nuclear genome (RST = 0.021, FST = 0.014; p = 0.308 ns).
Based on the global FST values derived from plastid haplotype data (FST (M) = 0.137), the global FSTs derived from nuclear microsatellite data (FST (B) = 0.018), and the inbreeding coefficient derived from nuclear microsatellite data (FIS(B) = 0.092; Table 3), the pollen to seed flow ratio in D. foliosa was calculated to mp/ms = 7.30.
Spatial autocorrelation calculations within geographic distances up to 256 m (Fig. 3) revealed a much steeper decline in pairwise kinskip, Fij, in the plastid genome than in the nuclear genome. For the plastid genome, kinship values were on average 0.528 in pairwise comparisons up to 4 m and were still significantly different from the permuted values at differences up to at least 256 m. For the nuclear genome, kinship coefficients were on average 0.133 in pairwise comparisons up to 4 m and were not significantly different from permuted values at distances above eight meters. The Sp statistic, describing the decline in average kinship values with distance, was estimated to Sp = 0.0264 for plastid haplotype data, and Sp = 0.0026 for the nuclear microsatellite data (Online resource 8).
Distogram reporting pairwise kinship estimates, Fij, between individuals of D. foliosa for distance intervals between 4 and 256 m. Separate calculations were made for plastid haplotype data (solid line) and nuclear microsatellite data (stippled line). To test for the hypotheses of significantly elevated levels of kinship within each distance category, the observed value was compared with the mean values obtained after 10,000 rounds of permutation of location of individuals among all locations. 95% confidence intervals for the permuted means are indicated by shading (light grey shading—plastid haplotypes; dark grey shading—nuclear microsatellites). One-hundred and seventy-three individual samples were examined
Discussion
Affinity to European members of Dactylorhiza
The molecular data obtained in the present study agree with a position of D. foliosa close to D. maculata subsp. maculata. All samples examined had a plastid haplotype corresponding to that of haplotype group II of Devos et al. (2003). Such haplotypes dominate in western European D. maculata subsp. maculata, but are more uncommon in D. maculata subsp. fuchsii (Ståhlberg and Hedrén 2010). In ITS, all plants of D. foliosa were fixed for the ITS allele designated as ITS-I in Pillon et al. (2007). The same ITS allele is strongly dominant in western European D. maculata subsp. maculata, but is rare in subsp. fuchsii (Pillon et al. 2007; Ståhlberg and Hedrén 2010). Of the four nuclear loci analysed, ms13 was fixed for an allele of 90 bp in all populations. This allele is also strongly dominant in western European D. maculata subsp. maculata and virtually absent from subsp. fuchsii (Hedrén and Ståhlberg, unpublished). Still, the diploid D. foliosa cannot be derived directly from the tetraploid D. maculata subsp. maculata, but rather from a common group of diploid ancestors to both of these taxa (Brandrud et al. 2020). Dactylorhiza foliosa also differs markedly from the European D. maculata subsp. maculata in general morphology, including the tall stature, the elongate and usually unspotted leaves and in the large pinkish flowers with indistinct markings. It is possible that these characteristics have evolved in relatively short time as a result of directional selection after the arrival of the ancestral population to Madeira. However, D. foliosa is also reminiscent of poorly studied members of the D. maculata group distributed in mountain regions of northwest Africa, including D. maculata subsp. battandieri and D. maculata subsp. maurusia (Landwehr 1977; Delforge 2001; Baumann et al. 2006). Further studies are obviously needed to understand the exact position of D. foliosa within the D. maculata s.l. complex.
Patterns of between-population differentiation in D. foliosa
A compilation of studies mostly based on allozyme data (Frankham 1997) has shown that island populations or island taxa have in most cases lower levels of genetic diversity than related continental counterparts. However, individual studies vary considerably in different directions, and some plants may have equally high amounts of gene diversity in island populations as in continental populations (e.g., García-Verdugo et al. 2015).
The levels of gene diversity observed in D. foliosa were comparable to levels found in other species of Dactylorhiza. The population average for nuclear markers, 0.453, was within the range observed in mainland taxa (0.355–0.668; Hedrén and Nordström Olofsson 2018; Hedrén et al. 2018, 2021). The gene diversity obtained for plastid haplotypes, 0.734, was even somewhat higher than that obtained in other taxa (0.259–0.675; op. cit.). Although estimated levels of genetic diversity depend on the degree of variability of the markers used in any particular study, the studies cited here for comparison with D. foliosa were based on partly the same markers, so the comparison should still be relevant.
The global FST calculated on basis of nuclear markers was 0.018 in D. foliosa. This value is lower than in most other orchids previously analysed for genetic diversity structure. The low FST value was also in agreement with the lack genetic structuring found by Bayesian clustering in Structure. Average estimates of GST/FST in orchids range from 0.087 (Hamrick and Godt 1996), over 0.146 (Phillips et al. 2012), to 0.187 (Forrest et al. 2004). Phillips et al. (2012) compared levels of FST for rare and common terrestrial orchids and found that the average FST was significantly lower in the latter category (FST = 0.279 vs FST = 0.092, respectively). Dactylorhiza foliosa is characterised as a species that is common within its range (Rankou 2011). Accordingly, the very low FST value and the absence of structuring in the nuclear genome could be an effect of the relatively high abundance and even distribution of D. foliosa in the laurel forest habitat on Madeira. The laurel forest is more or less continuous on the northern side of the island (Fig. 1), and there appears to be no major barriers to dispersal within the distribution range.
It should also be observed that the total area occupied by D. foliosa is small. The sampled populations were all relatively close to each other and are probably more efficiently connected by gene flow than populations of other studied orchid species, which have larger and patchier distributions.
A third factor likely to contribute to a low FST in D. foliosa is the pollination system. Like other members of Dactylorhiza, D. foliosa is characterized by a deceptive pollination system (Baumann et al. 2006) and it offers no nectar to visiting insects. Although rewarding orchids have higher fruit set than orchids offering no reward to their pollinators (Gill 1989, Neiland and Wilcock 1995), it has been suggested that the pollinators of non-rewarding orchids ensure more efficient cross-pollination as they transport pollen longer distances between plants (Nilsson 1992, Neiland and Wilcock 1995, Jersáková et al., 2006). Empirical data show that orchids characterized by deceptive pollination systems have on average a significantly lower level of population differentiation than orchids offering nectar or other rewards to their pollinators (FST = 0.25 vs FST = ca 0.13; Cozzolino and Widmer 2005).
Many orchid species suffer from inbreeding depression following selfing (Smithson 2006). Only a small proportion of orchids are autogamous or asexual. Inbreeding levels are often moderate in orchids, but fixation indices, FIS, may range from 0 to 1. Inbreeding levels in other members of Dactylorhiza range from FIS = 0.15–0.66 (Hedrén & Lorenz 2019). The estimated level of inbreeding was moderate in D. foliosa with mean FIS = 0.222 (Table 1), and somewhat lower still if the two smallest population samples were excluded. As the two smallest populations did hardly differ from other sampled populations in plant density (online resource 1), it is possible that the small number of potential mates, rather than the mean distance between plants, provides the most likely explanation to the observed variation in inbreeding levels.
The populations of D. foliosa were more strongly differentiated in plastid haplotypes than in nuclear microsatellites (FST = 0.137). However, this value is also quite low in comparison with other orchid species, including members of Dactylorhiza in which it ranges from 0.258 to 0.707 (Hedrén et al. 2021).
Since the plastid genome is exclusively dispersed by seeds in orchids (Corriveau and Coleman 1988), whereas the nuclear genome is dispersed by both seeds and pollen, FST estimates based on plastid data are expected to be consistently higher than FSTs based on nuclear markers (Petit et al. 2005). Still, orchids have minute seeds that are supposedly easily dispersed long distances by wind (Arditti and Ghani 2000), and the seeds might contribute disproportionally much to gene dispersal, as compared to other plant families with heavier seeds.
The two FST measures can be combined with the inbreeding coefficient, FIS, to calculate the pollen to seed flow ratio of Ennos (1994), also given as mp/ms in Petit et al. (2005). Whereas orchids often have low values for this ratio, sometimes close to one (Squirrel et al. 2001; Cozzolino et al. 2003), Petit et. al. (2005) calculated a median value of mp/ms across all investigated seed plants as mp/ms = 17.
The pollen to seed flow ratio in D. foliosa was calculated to mp/ms = 7.32. This ratio is slightly higher than in estimates obtained from other members of Dactylorhiza investigated so far (Hedrén et al. 2021). However, the pollen to seed flow ratio might be dependent on geographic scale (McCauley 1997; Hamrick and Trapnell 2011; Hedrén et al. 2021). Whereas pollen dispersal typically declines gradually from the pollen source, depending on the flight patterns of the pollinators, seed dispersal distances typically exhibit a strongly leptokurtic distribution, with large amounts of seeds deposited in the close vicinity of the source plant and only a minor portion dispersed at medium and long distances (Cain et al. 1998). In effect, the relative contribution of pollen dispersal to total gene dispersal is small at short distances, higher within populations and between close populations, but again small as compared to seed dispersal at long distances (McCauley 1997; Hamrick and Trapnell 2011). For example, in the hummingbird-pollinated epiphytic orchid Laelia rubescens in Costa Rica, mp/ms ratios up to 13.67 were recorded for populations separated by less than 2 km, but a ratio of 1.40 between populations at a scale up to 10 km (Trapnell and Hamrick 2004), Likewise, in the terrestrial, bumble bee-pollinated Dactylorhiza umbrosa investigated on the Anatolian-Iranian highland plateau, mp/ms ratios of 8.4 – 12.01 were recorded at distances up to 122 km, but ratios of 2.8–4.9 at higher distances (Hedrén et al. 2021). It is possible therefore, that the high value recorded for D. foliosa, with a maximum distance between populations of 26 km, reflects the relatively small sampling area.
Alternatively, the comparatively high pollen to seed flow ratio recorded for D. foliosa could be a consequence of the sheltered forest habitat in which the species is usually found. A forest acts as shelter to strong winds, especially at ground level, and might thus reduce dispersal distances of small, wind-borne seeds. In contrast, as pollen is dispersed by actively flying insects, pollen dispersal in a forest habitat might not be as strongly affected. Other species of Dactylorhiza that have been examined for genetic diversity structure are typically found in wet grasslands or in open fen habitats where seeds are more efficiently dispersed by winds.
There was no significant correlation between pairwise FSTs derived from plastid data and geographic distance between populations, and likewise, there was no significant correlation between pairwise FSTs derived from nuclear data and geographic distance (Table 2). Although the amount of population differentiation as measured by FST is generally low in orchids, Phillips et al. (2012) found that many common orchids are still characterized by an isolation by distance structure; IBD. However, they also found that IBD was most often detected in studies where the scale of sampling exceeded 250 km, far longer than the 26 km separating the most distant populations sampled of D. foliosa.
I also tested each of the plastid and nuclear microsatellite data sets for the presence of a phylogeographic structure, such that divergent haplotypes or microsatellite alleles in different population are in general more strongly differentiated than divergent haplotypes or alleles present in the same population (Hardy and Vekemans 2002). In D. foliosa there was no evidence for any significant phylogeographic structure in the nuclear microsatellite data, but for the plastid data, NST, which takes numbers of differences between haplotypes into account, was significantly higher than the permuted value corresponding to an ordinary GST (0.400 vs. 0.229, p < 0.001***). An indication of a phylogeographic structure in the plastid genome is also given by the parsimony network (Fig. 3) in which it could be observed that geographically adjacent sampling sites often share the same or related plastid haplotypes with each other; viz. the three subsites at Encumeada, the close sites Quiemadas and Pico das Pedras, and the two easternmost sampling sites Ribeiro Frio and Portela. However, there is also cases where distant populations share the same haplotypes. Such cases might be explained by rare long-distance dispersal events, or simply homoplasy in the data. The interpretation of the combined patterns is that gene dispersal by seeds is not efficient enough to balance out the differentiation that slowly accumulates as result of local mutations within populations of D. foliosa. In contrast, pollen dispersal seems to be effective to counterbalance differentiation by mutation in nuclear microsatellites.
Spatial genetic structure
The genetic differentiation patterns within sampling sites was analysed by spatial autocorrelation and by comparison of mean values of kinship, Fij, for different distance intervals at distances up to 256 m (Fig. 3, Online resource 6). Whereas plants close to each other often shared the same plastid haplotypes and still did so significantly more often at distances up to at least 256 m, the kinship estimate derived from nuclear microsatellites was small already at close distance and kinship values were not significantly different from permuted values at distances above eight metres. Accordingly, pollen dispersal contributes much more to total gene dispersal than seed dispersal also at the local scale.
The Sp value calculated for the nuclear data (Sp = 0.0026) was very low in comparison with most other orchids examined (range 0.0015–0.069 in predominantly outbreeding species; Chung et al. 2011; Hedrén and Lorenz 2019). However, the value found in D. foliosa was still comparable to some populations of the Asian terrestrial species Cymbidium goeringii (Chung and Nason 2007).
The Sp value calculated on basis of plastid haplotype data was 0.0264 in D. foliosa. There are no other studies in which Sp statistics for the plastid genome has been reported for an orchid species. However, in some orchids characterized by uniparental reproduction, and in which pollen dispersal does not contribute to gene dispersal, Sp values have been found to be higher than in the plastid genome in D. foliosa, with values ranging from 0.05 to 0.208 (Hedrén and Lorenz 2019).
Conclusions
The amount and structuring of genetic diversity in D. foliosa were comparable to members of Dactylorhiza growing in continental areas. Populations of D. foliosa were also not strongly differentiated from each other and gene dispersal within the species–especially gene dispersal by pollen–seems to connect populations across the species range. A stronger differentiation between populations was observed in plastid haplotypes, and the plastid data expressed a significant phylogeographic structure, pointing towards a somewhat restricted seed dispersal. On basis of molecular marker data, it appears that D. foliosa contains high enough levels of genetic diversity to support long-term persistence and future adaptation on Madeira. However, care should be taken to preserve the laurel forest habitat on the island, to maintain connectivity across the species range and to maintain sufficiently large populations. Fragmentation of the habitat might result in reduction in gene flow, smaller population sizes and increased levels of inbreeding, which in turn might lead to loss of genetic diversity and an increased risk of population loss and extinction.
Availability of data and material
All data generated and analysed during this study are included in this published article [and its supplementary information files].
References
Arditti J, Ghani AKA (2000) Numerical and physical properties of orchid seeds and their biological implications. New Phytol 145:367–421
Bateman RM, Hollingsworth PM, Preston J, Luo Y-B, Pridgeon AM, Chase MW (2003) Molecular phylogenetics and evolution of Orchidinae and selected Habenariinae (Orchidaceae). Bot J Linn Soc 142:1–40. https://doi.org/10.1046/j.1095-8339.2003.00157.x
Baumann H, Künkele S, Lorenz R (2006) Orchideen Europas. Eugen Ulmer, Stuttgart
Brandrud MK, Baar J, Lorenzo MT, Bateman RM, Chase MW, Hedrén M, Paun O (2020) Phylogenomic relationships of diploids and the origins of allotetraploids in Dactylorhiza (Orchidaceae): RADseq data track reticulate evolution. Systamatic Biology 69:91–109. https://doi.org/10.1093/sysbio/syz035
Cain ML, Damman H, Muir A (1998) Seed dispersal and the holocene migration of woodland herbs. Ecol Monogr 68:325–347
Capelo J, Sequeira M, Jardim R, Mesquita S, Costa JC (2005) The vegetation of Madeira Island (Portugal). A brief overview and excursion guide. Quercetea 7:95–122
Chase MW, Hills HG (1991) Silica gel: an ideal material for field preservation of leaf samples for DNA studies. Taxon 40:215–220
Chung MY, Nason JD (2007) Spatial demographic and genetic consequences of harvesting within populations of the terrestrial orchid Cymbidium goeringii. Biol Conservation 137:125–137. https://doi.org/10.1016/j.biocon.2007.01.021
Chung MY, Nason JD, Chung MG (2011) Significant demographic and fine-scale genetic structure in expanding and senescing populations of the terrestrial orchid Cymbidium goeringii (Orchidaceae). Amer J Bot 98:2027–2039. https://doi.org/10.3732/ajb.1100094
Corriveau JL, Coleman AW (1988) Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. Amer J Bot 75:1443–1458
Cox CB, Healey IN, Moore PD (1976) Biogeography. Blackwell, Oxford UK
Cozzolino S, Widmer A (2005) Orchid diversity: an evolutionary consequence of deception? Trends Ecol Evol 20:487–494. https://doi.org/10.1016/j.tree.2005.06.004
Cozzolino S, Cafasso D, Pellegrino G, Musacchio A, Widmer A (2003) Fine-scale phylogeographical analysis of Mediterranean Anacamptis palustris (Orchidaceae) populations based on chloroplast minisatellite and microsatellite variation. Molec Ecol 12:2783–2792. https://doi.org/10.1046/j.1365-294X.2003.01958.x
Delforge P (2001) Guide des orchidées d´Europe, d´Afrique du nord et du proche-orient, 2nd edn. Delachaux et Niestlé, Lausanne
Devos N, Tyteca D, Raspé O, Wesselingh RA, Jacquemart A-L (2003) Patterns of chloroplast diversity among western European Dactylorhiza species (Orchidaceae). Pl Syst Evol 243:85–97. https://doi.org/10.1007/s00606-003-0068-0
Devos N, Oh S-H, Raspé O, Jacquemart A-L, Manos PS (2005) Nuclear ribosomal DNA sequence variation and evolution of spotted marsh-orchids (Dactylorhiza maculata group). Molec Phylogen Evol 36:568–580. https://doi.org/10.1016/j.ympev.2005.04.014
Doyle JJ, Doyle JH (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Earl DA, vonHoldt BM (2012) Structure harvester: a website and program for visualizing structure output and implementing the Evanno method. Conservation Genet Resources 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Ennos RA (1994) Estimating the relative rates of pollen and seed migration among plant populations. Heredity 72:250–259
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Molec Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Fernandes FM, Carvalho MÂAP, Aguiar AF (2005) Pollination of madeiran indigenous orchids. In: Carvalho MÂAP et al (eds) Proceedings of the II symposium island ecosystems centre for macaronesian studies. University of Madeira, Madeira, pp 25–36
Fluxus technology (2007) Network 4.2.0.1. User Guide. Available at: http://www.fluxux-technology.com
Forrest AD, Hollingsworth ML, Hollingsworth PM, Sydes C, Bateman RM (2004) Population genetic structure in European populations of Spiranthes romanzoffiana set in the context of other genetic studies on orchids. Heredity 92:218–227. https://doi.org/10.1038/sj.hdy.6800399
Frankel OH, Soulé ME (1981) Conservation and evolution. Cambridge University Press, Cambridge UK
Frankham R (1997) Do island populations have less genetic variation than mainland populations? Heredity 78:311–327
García-Verdugo C, Sajeva M, La Mantia T, Harrouni C, Msanda F, Caujapé-Castells J (2015) Do island plant populations really have lower genetic variation than mainland populations? Effects of selection and distribution range on genetic diversity estimates. Molec Ecol 24:726–741. https://doi.org/10.1111/mec.13060
Gill DE (1989) Fruiting failure, pollinator inefficiency, and speciation in orchids. In: Otte D, Endler JA (eds) Speciation and its consequences. Sinauer, Sunderland, pp 458–481
Hamrick JL, Godt MJW (1990) Allozyme diversity in plant species. In: Brown ADH, Clegg MT, Kahler AH, Weir BS (eds) Plant population genetics, breeding and genetic resources. Sinauer, Sunderland, pp 43–63
Hamrick JL, Godt MJW (1996) Effects of life history traits on genetic diversity in plant species. Phil Trans Roy Soc London Ser B Biol Sci 351:1291–1298
Hamrick JL, Trapnell DW (2011) Using population genetic analyses to understand seed dispersal patterns. Acta Oecol 37:641–649. https://doi.org/10.1016/j.actao.2011.05.008
Hardy OJ, Vekemans X (2002) Spagedi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molec Ecol Notes 2:618–620. https://doi.org/10.1046/j.1471-8278.2002.00305.x
Hedrén M, Lorenz R (2019) Seed dispersal and fine-scale genetic structuring in the asexual Nigritella miniata (Orchidaceae) in the Alps. Bot J Linn Soc 190:83–100. https://doi.org/10.1093/botlinnean/boz005
Hedrén M, Nordström Olofsson S (2018) High levels of genetic diversity in marginal populations of the marsh orchid Dactylorhiza majalis ssp. majalis. Nordic J Bot 36:njb-01747. https://doi.org/10.1111/njb.01747
Hedrén M, Fay M, Chase MW (2001) Amplified fragment length polymorphisms (AFLP) reveal details of polyploid evolution in Dactylorhiza (Orchidaceae). Amer J Bot 88:1868–1880
Hedrén M, Nordström S, Ståhlberg D (2008) Polyploid evolution and plastid DNA variation in the Dactylorhiza incarnata/maculata complex (Orchidaceae) in Scandinavia. Molec Ecol 17:5075–5091. https://doi.org/10.1111/j.1365-294X.2008.03965.x
Hedrén M, Nordström Olofsson S, Paun O (2018) Orchid colonization: multiple parallel dispersal events and mosaic genetic structure in Dactylorhiza majalis ssp. lapponica on the Baltic island of Gotland. Ann Bot (Oxford) 122:1019–1031. https://doi.org/10.1093/aob/mcy111
Hedrén M, Birkedal S, de Boer H, Ghorbani A, Gravendeel B, Hansson S, Svensson Å, Zarre S (2021) Asymmetric contributions of seed and pollen to gene dispersal in the marsh orchid Dactylorhiza umbrosa in Asia Minor. Molec Ecol 30:1791–1805. https://doi.org/10.1111/mec.15848
Irl SDH, Schweiger AH, Medina FM, Fernández-Palacios JM, Harter DEV, Jentsch A, Provenzale A, Steinbauer MJ, Beierkuhnlein C (2017) An island view of endemic rarity – Environmental drivers and consequences for nature conservation. Diversity Distrib 23:1132–1142. https://doi.org/10.1111/ddi.12605
Jakobsson M, Rosenberg NA (2007) Clumpp: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806. https://doi.org/10.1093/bioinformatics/btm233
Jardim R, Francisco D (2000) Flora Endémica da Madeira. Múchia Publicações, Funchal
Jersáková J, Johnson SD, Kindlmann P (2006) Mechanisms and evolution of deceptive pollination in orchids. Biol Rev 81:219–236. https://doi.org/10.1017/S1464793105006986
Kier G, Kreft H, Lee TM, Jetz W, Ibisch PL, Nowicki C, Mutke J (2009) A global assessment of endemism and species richness across island and mainland regions. Proc Nat Acad Sci USA 106:9322–9327. https://doi.org/10.1073/pnas.0810306106
Lande R (1999) Extinction risks from anthropological, ecological and genetic factors. In: Landweber LF, Dodson AP (eds) Genetics and the extinction of species. Princeton University Press, Princeton, pp 1–22
Landwehr J (1977) Wilde orchideeên van Europa, vol. 1. Vereiniging tot behoud van natuurmonumenten in Nederland, 's-Graveland
Loiselle BA, Sork VL, Nason JD, Graham C (1995) Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Amer J Bot 82:1420–1425
McCauley DE (1997) The relative contributions of seed and pollen movement to the local genetic structure of Silene alba. J Heredity 1997:257–263
Michalakis Y, Excoffier L (1996) A genetic estimation of population subdivision using distances between alleles with special reference for microsatellite loci. Genetics 142:1061–1064
Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858
Neiland MRM, Wilcock CC (1995) Maximisation of reproductive success by European Orchidaceae under conditions of infrequent pollination. Protoplasma 187:39–48
Nilsson LA (1992) Orchid pollination biology. Trends Ecol Evol 7:225–259
Nordström S, Hedrén M (2007) Development of polymorphic nuclear microsatellite markers for polyploid and diploid members of the orchid genus Dactylorhiza. Molec Ecol Notes 7:644–647. https://doi.org/10.1111/j.1471-8286.2006.01662.x
Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin GG (2005) Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Molec Ecol 14:689–701. https://doi.org/10.1111/j.1365-294X.2004.02410.x
Phillips R, Dixon K, Peakall R (2012) Low population genetic differentiation in the Orchidaceae: implications for the diversification of the family. Molec Ecol 21:5208–5220. https://doi.org/10.1111/mec.12036
Pillon Y, Fay MF, Hedrén M, Bateman RM, Devey D, Shipunov A, van der Bank M, Chase MW (2007) Insights into the evolution and biogeography of Western European species complexes in Dactylorhiza (Orchidaceae). Taxon 56:11851208
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rankou H (2011) Dactylorhiza foliosa. The IUCN Red list of threatened species 2011: e.T176013A7171459. Available at: https://doi.org/10.2305/IUCN.UK.2011-2.RLTS.T176013A7171459.en. Accessed 6 May 2016
Rohlf FJ (2005) NTSYSpc: numerical taxonomy system, 2 20. Exeter Publishing, Setauket
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Slatkin M (1995) Gene flow in natural populations. Annual Rev Ecol Syst 16:393–430
Smithson A (2006) Pollinator limitation and inbreeding depression in orchid species with and without nectar rewards. New Phytol 169:419–430. https://doi.org/10.1111/j.1469-8137.2005.01592.x
Squirrel J, Hollingsworth PM, Bateman RM, Dickson JH, Light MHS, MacConaill M, Tebbitt MC (2001) Partitioning and diversity of nuclear and organelle markers in native and introduced populations of Epipactis helleborine (Orchidaceae). Amer J Bot 88:1409–1418
Ståhlberg D, Hedrén M (2010) Evolutionary history of the Dactylorhiza maculata polyploid complex (Orchidaceae). Biol J Lin Soc 101:503–525
Trapnell DW, Hamrick JL (2004) Partitioning nuclear and chloroplast variation at multiple spatial scales in the neotropical epiphytic orchid, Laelia rubescens. Molec Ecol 13:2655–2666. https://doi.org/10.1111/j.1365-294X.2004.02281.x
Vekemans X, Hardy OJ (2004) New insights from fine-scale genetic structure analyses in plant populations. Molec Ecol 13:921–935. https://doi.org/10.1046/j.1365-294X.2004.02076.x
Veron S, Haevermans T, Govaerts R, Mouchet M, Pellens R (2019) Distribution and relative age of endemism across islands worldwide. Sci Rep 11:693. https://doi.org/10.1038/s41598-019-47951-6
Weir BS, Cockerham CC (1984) Estimating F-Statistics for the analysis of population structure. Evolution 38:1358–1370
Wright S (1943) Isolation by distance. Genetics 28:114–138
Acknowledgements
I thank Richard Bateman and one anonymous reviewer for helpful comments on the manuscript. The study was supported by grants from Lunds Botaniska Förening and Elly Olssons Fond.
Funding
Open access funding provided by Lund University. This study was supported by grants from Lunds Botaniska Förening and Elly Olssons Fond.
Author information
Authors and Affiliations
Contributions
The author designed the study, sampled the material, conducted lab analyses, performed statistical analyses, and wrote the manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The author declares no competing interests.
Additional information
Handling Editor: Andreas Tribsch.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online Resource 1. Sampling locations of Dactylorhiza foliosa.
Online Resource 2. Overview of molecular markers studied in Dactylorhiza foliosa.
Online Resource 3. Haplotypes in Dactylorhiza foliosa separated on basis of combined variation at four plastid marker loci.
Online Resource 4. Base data.
Online Resource 5. Allele frequencies per population at nuclear microsatellite loci in Dactylorhiza foliosa.
Online Resource 6. Examples of bar plots obtained from running the computer program Structure 2.3.4 (Pritchard et al. 2000).
Online Resource 7. Delta K values for K=2 to K=9 in the material studied of Dactylorhiza foliosa as estimated by the Evanno method (Evanno et al. 2005) and calculated in the computer program Structure Harvester (Earl and vonHoldt 2012).
Online Resource 8. Fine-scale spatial genetic structure in Dactylorhiza foliosa. One-hundred and seventy-three individual samples were examined. Separate analyses were performed for nuclear data (three variable microsatellite loci) and plastid data (plastid haplotypes) Pairwise kinship coefficients, Fij, were calculated according to Loiselle et al. (1995).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hedren, M. Spatial genetic structure in the Madeiran endemic Dactylorhiza foliosa (Orchidaceae). Plant Syst Evol 308, 30 (2022). https://doi.org/10.1007/s00606-022-01822-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00606-022-01822-2