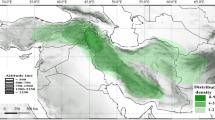
Abstract
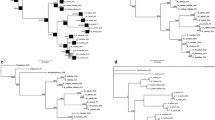
The present study was conducted to analyze the phylogenetic status of the genus Onobrychis and to evaluate the monophyly of its subgenera and sections and relationship among them. We sequenced the nuclear ribosomal DNA internal transcribed spacer (nrDNA ITS) and three chloroplast regions trnL-F, rpl32/rpl32-trnL(UAG) and ndhF-rlp32 for phylogenetic reconstruction of 51 species of Onobrychis. In all of our analyses, Eversmannia subspinosa, Corethrodendron scoparium, Greuteria membranacea and G. argyrea were chosen as outgroups. Phylogenetic analyses were performed by maximum parsimony, maximum likelihood and Bayesian methods. Our molecular data indicate that Onobrychis is monophyletic and composed of two main clades, each corresponding to the redefined subgenus Onobrychis (including sections Onobrychis and Hemicyclobrychis) and subgenus Sisyrosema (including sections Afghanicae, Laxiflorae, Heliobrychis, Hymenobrychis, Insignes, Lipskyanae and Litvinovianae), respectively. Sections Lipskyanae and Litvinovianae are newly established and described, representing distinct lineages within the genus. Onobrychis splendida, a species hitherto without a sectional position, along with some members of sect. Anthyllium were retrieved representatives of section Lipskyanae. Sections Afghanicae, Insignes, Heliobrychis and Hymenobrychis (with the inclusion of two species of section Anthyllium) are monophyletic. Sections Dendrobrychis and Lophobrychis are reduced to synonymy of section Onobrychis and Anthyllium to synonymy of section Hymenobrychis. A taxonomic treatment for the genus is presented.

Similar content being viewed by others
References
Abou-El-Enain M (2002) Chromosomal criteria and their phylogenetic implications in the genus Onobrychis Mill. sect. Lophobrychis (Leguminosae), with special reference to Egyptian species. Bot J Linn Soc 139:409–414. doi:10.1046/j.1095-8339.2002.00075.x
Ahangarian S, Kazempour Osaloo S, Maassoumi AA (2007) Molecular phylogeny of the tribe Hedysareae with special reference to Onobrychis (Fabaceae) as inferred from nrDNA ITS sequences. Iranian J Bot 13:64–74
Amirabadizadeh H, Abbassi M, Ranjbar M (2007) A new species of Onobrychis Sect. Heliobrychis (tribe Hedysarae) from Iran. Iranian J Bot 13:53–56
Amirabadizadeh H, Ghanavati F, Abbassi M, Ranjbar M (2009) A new species of Onobrychis sect. Afghanicae (Fabaceae) from Iran. Iranian J Bot 15:45–50
Amirahmadi A, Kazempour Osaloo S, Moein F, Kaveh A, Maassoumi AA (2014a) Molecular systematics of the tribe Hedysareae (Fabaceae) based on nrDNA ITS and plastid trnL-F and matK sequences. Pl Syst Evol 300:729–747. doi:10.1007/s00606-013-0916-5
Amirahmadi A, Kazempour Osaloo S, Khoshsokhan-Mozaffar M, Charkhchian MM (2014b) A new species of Onobrychis sect. Onobrychis (Fabaceae) from Iran. Turkish J Bot 38:658–664. doi:10.3906/bot-1309-54
Arslan E, Ertuğrul K, Tugay O, Dural H (2012) Karyological studies of the genus Onobrychis Mill. and the related genera Hedysarum L. and Sartoria Boiss. & Heldr. (Fabaceae, Hedysareae) from Turkey. Caryologia 65:11–17. doi:10.1080/00087114.2012.678079
Avci S, Sancak C, Can A, Acar A, Pınar NM (2013) Pollen morphology of the genus Onobrychis (Fabaceae) in Turkey. Turkish J Bot 37:669–681. doi:10.3906/bot-1207-52
Baldwin BJ, Sanderson MJ, Porter MJ, Wojciechowski MF, Campbell C, Donoghue JM (1995) The ITS region of nuclear ribosomal DNA: a valuable source of evidence of Angiosperm phylogeny. Ann Missouri Bot Gard 82:247–277. doi:10.2307/2399880
Ball PW (1978) Onobrychis. In: Tutin TG, Heywood VH, Burges NA, Moore DM, Valentine DH, Walters SM, Webb DA (eds) Flora Europaea, vol 2. Cambridge University Press, Cambridge, pp 187–191
Cunningham CW (1997) Can three incongruence tests predict when data should be combined? Molec Biol Evol 14:733–740
Dong W, Liu J, Yu J, Wang L, Zhou S (2012) Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE 7:e35071. doi:10.1371/journal.pone.0035071
Douzery E, Pridgeon A, Kores P, Linder HP, Kurzweil H, Chase M (1999) Molecular phylogenetics of Diseae (Orchidaceae): a contribution from nuclear ribosomal ITS sequences. Amer J Bot 86:887–899
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Duan L, Wen J, Yang X, Liu PL, Arslan E, Ertuğrul K, Chang ZY (2015) Phylogeny of Hedysarum and tribe Hedysareae (Leguminosae: Papilionoideae) inferred from sequence data of ITS, matK, trnL-F and psbA-trnH. Taxon 64:49–64. doi:10.12705/641.26
Edgar RC (2004) Muscle: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res 32:1792–1797. doi:10.1093/nar/gkh340
Farris JS, Kallersjo M, Kluge AG, Bult C (1995) Testing significance of incongruence. Cladistics 10:315–319. doi:10.1111/j.1096-0031.1994.tb00181.x
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 38:783–791. doi:10.2307/2408678
Grossheim AA (1972) Onobrychis Adans. (Leguminosae). In: Komarov VL, Shishkin BK, Bobrov EG (eds) Flora of the USSR, vol 13. Israel Program for Scientific Translations, Jerusalem, pp 244–281
Hayot Carbonero C, Carbonero F, Smith LMJ, Brown TA (2012) Phylogenetic characterization of Onobrychis species with special focus on the forage crop Onobrychis viciifolia Scop. Genet Resources Crop Evol 59:1777–1788. doi:10.1007/s10722-012-9800-3
Hedge IC (1970) Onobrychis (Leguminosae-Hedysareae). In: Davis PH, Chamberlain DF, Matthews VA (eds) Flora of Turkey and the East Aegean Islands, vol 3. Edinburgh University Press, Edinburgh, pp 560–590
Hesamzadeh Hejazi SM, Ziaei Nasab M (2010) Cytotaxonomy of some Onobrychis (Fabaceae) species and populations in Iran. Caryologia 63:18–31. doi:10.1080/00087114.2010.589705
Irfan E, Turgut-Balik D, Sahin A, Kursat M (2007) Total electrophoretic band patterns of some Onobrychis species growing in Turkey. Amer Eurasian J Agric Environ Sci 2:123–126
Karamian R, Moradi Behjou A, Ranjbar M (2012) Anatomical findings of Onobrychis sect. Heliobrychis (Fabaceae) in Iran and their taxonomic implications. Turkish J Bot 36:27–31. doi:10.3906/bot-1010-2
Kazempour Osaloo S, Maassoumi AA, Murakami N (2005) Molecular systematics of the Old World Astragalus (Fabaceae) as inferred from nrDNA ITS sequence data. Brittonia 57:367–381. doi:10.1663/0007-196X(2005)057[0367:MSOTOW]2.0.CO;2
Lamarck MM, De Candolle AP (1805) Flore Française. vol. 4, part 2, Paris, pp 401–946
Lewke Bandara N, Papini A, Mosti S, Brown T, Smith LMJ (2013) A phylogenetic analysis of genus Onobrychis and its relationships within the tribe Hedysareae (Fabaceae). Turkish J Bot 37:981–992. doi:10.3906/bot-1210-32
Lock JM (2005) Tribe Hedysarae. In: Lewis G, Schrire B, Mackinder B, Lock M (eds) Legumes of the world. Royal Botanical Gardens, Kew, pp 489–495
Mabberley DJ (2008) The plant-book. A portable dictionary of the higher plants, 3rd edn. Cambridge University Press, Cambridge
Nylander JAA (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Uppsala
Page DM (2001) Treeview (Win32) version 1.6.6. Available at: http://taxonomy.zoology.gla.ac.uk/rod/treeview.html
Pavlova DK, Manova VI (2000) Pollen morphology of the genera Onobrychis and Hedysarum (Hedysareae, Fabaceae) in Bulgaria. Ann Bot Fenn 37:207–217
Podlech D (1967) Neue und bemerkenswerte Fabaceae aus Nordost-Afghanistan (beiträge zur Flora von Afghanistan II). Mitt Bot Staatssamml München 6:547–591
Posada D, Buckley TR (2004) Model selection and model averaging in phylogenetics: advantages of akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol 53:793–808. doi:10.1080/10635150490522304
Ranjbar M, Amirabadizadeh H, Karamian R, Ghahremani MA (2004) Notes on Onobrychis sect. Heliobrychis (Fabaceae) in Iran. Willdenowia 34:187–190. doi:10.3372/wi.34.34116
Ranjbar M, Karamian R, Hajmoradi F (2009) Taxonomic notes on Onobrychis sect. Hymenobrychis (Fabaceae, Hedysareae) in Iran. Novon 19:215–218. doi:10.3417/2007119
Ranjbar M, Karamian R, Hadadi A (2010) Cytosystematics of three Onobrychis species (Fabaceae) in Iran. Caryologia 63:237–249. doi:10.1080/00087114.2010.10589733
Ranjbar M, Hajmoradi F, Karamian R (2012) An overview on cytogenetics of the genus Onobrychis (Fabaceae) with special reference to O. sect. Hymenobrychis from Iran. Caryologia 65:187–198. doi:10.1080/00087114.2012.735887
Rechinger KH (1984) Hedysareae. In: Rechinger KH (ed) Flora Iranica, vol 157. Akademische Druck, Graz, pp 365–475
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi:10.1093/sysbio/sys029
Safaei Chaei Kar S, Ghanavati F, Mozafari J, Naghavi MR, Amirabadizadeh H, Darvish F (2012) Phylogenetic relationships of Onobrychis Mill. (Fabaceae: Papilionoideae) based on ITS sequences of nuclear ribosomal DNA and morphological traits. Crop Breed J 2:91–99
Safaei Chaei Kar S, Ghanavati F, Naghavi MR, Amirabadi-zade H, Rabiee R (2014) Molecular phylogenetics of the Onobrychis genus (Fabaceae: Papilionoideae) using ITS and trnL–trnF DNA sequence data. Austral J Bot 62:235–250. doi:10.1071/BT13279
Sang T, Crawford DJ, Stuessy T (1995) Documentation of reticulate evolution in peonies (Paeonia) using internal transcribed spacer sequences of nuclear ribosomal DNA: implication for biogeography and concerted evolution. Proc Natl Acad Sci USA 92:6813–6817
Shaw J, Lickey EB, Schilling EE, Small RL (2007) Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in Angiosperms: the tortoise and the hare III. Amer J Bot 94:275–288. doi:10.3732/ajb.94.3.275
Silvestro D, Michalak I (2012) raxmlGUI: a graphical front-end for RAxML. Organisms Divers Evol 12:335–337. doi:10.1007/s13127-011-0056-0
Širjaev G (1925) Onobrychis generis revisio critica, pars prima. Spisy Přír Fak Masarykovy Univ 56:1–197
Širjaev G (1926) Onobrychis generis revisio critica, partes secunda et tertia. Spisy Přír Fak Masarykovy Univ 76:1–165
Swofford DL (2002) PAUP*: Phylogenetic analysis using parsimony (*and other methods), Version 4.0b10. Sinauer Associates, Sunderland
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal Primers for amplification of three non-coding regions of chloroplast DNA. Pl Molec Biol 17:1105–1109. doi:10.1007/BF00037152
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Molec Biol Evol 30:2725–2729. doi:10.1093/molbev/mst197
Townsend CC (1974) Papilionaceae. In: Townsend CC, Guest ER (eds) Flora of Iraq, vol 3. Ministry of Agriculture and Agrarian Reform of the Republic of Iraq, Baghdad, pp 54-601
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis DH (ed) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
Wojciechowski MF, Sanderson MJ, Hu JM (1999) Evidence on the monophyly of Astragalus (Fabaceae) and its major subgroups based on nuclear ribosomal DNA ITS and chloroplast DNA trnL intron data. Syst Bot 24:409–437. doi:10.2307/2419698
Yildiz B, Ciplak B, Aktoklu E (1999) Fruit morphology of sections of the genus Onobrychis Miller (Fabaceae) and its phylogenetic implications. Israel J Pl Sci 47:269–282. doi:10.1080/07929978.1999.10676784
Zarrabian M, Majidi MM (2015) Genetic diversity and relationships within and among Onobrychis species using molecular markers. Turkish J Bot 39:681–692. doi:10.3906/bot-1406-47
Zwickl D (2006) Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD thesis, University of Texas, Austin
Acknowledgments
The present study was financially supported in part by Grant-in-Aids for Scientific Research, No. 89002433, to S.K. O (corresponding author) from INSF (Iran National Science Foundation). We would like to thank the staff of the herbaria of, FUMH, HKNRRC, HQNRRC, MSB, TARI and TUH to allow studying herbarium specimens and providing leaf materials. We also thank H. Zare for editing and improving linguistic of the text. The trees resulting from Parsimony, Likelihood and Bayesian analyses of nrDNA ITS and plastid datasets, trnL–F, rpl32/rpl32-trnL(UAG) and ndhF-rpl32 are presented as Electronic Supplementary Material in Online resource 1, 2, 3, 4. The aligned data matrix used in this study is presented as Online Resource 5.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: Christoph Oberprieler.
Electronic supplementary material
Below is the link to the electronic supplementary material.
606_2016_1343_MOESM1_ESM.pdf
Online Resource 1 Fifty percent majority rule consensus tree resulting from Bayesian inference of the nrDNA ITS dataset. Numbers above branches are posterior probability and likelihood as well as parsimony bootstrap values, respectively. Values<50 % were not shown (PDF 132 kb)
606_2016_1343_MOESM2_ESM.pdf
Online Resource 2 Fifty percent majority rule consensus tree resulting from Bayesian inference of the plastid trnL-F dataset. Numbers above branches are posterior probability and likelihood as well as parsimony bootstrap values, respectively. Values<50 % were not shown (PDF 129 kb)
606_2016_1343_MOESM3_ESM.pdf
Online Resource 3 Fifty percent majority rule consensus tree resulting from Bayesian inference of the plastid rpl32/rpl32- trnLUAG dataset. Numbers above branches are posterior probability and likelihood as well as parsimony bootstrap values, respectively. Values<50 % were not shown (PDF 129 kb)
606_2016_1343_MOESM4_ESM.pdf
Online Resource 4 Fifty percent majority rule consensus tree resulting from Bayesian inference of the plastid ndhF-rpl32 dataset. Numbers above branches are posterior probability and likelihood as well as parsimony bootstrap values, respectively. Values<50 % were not shown (PDF 132 kb)
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online Resource 1. Fifty percent majority rule consensus tree resulting from Bayesian inference of the nrDNA ITS dataset.
Online Resource 2. Fifty percent majority rule consensus tree resulting from Bayesian inference of the plastid trnL-F dataset.
Online Resource 3. Fifty percent majority rule consensus tree resulting from Bayesian inference of the plastid rpl32/rpl32- trnLUAG dataset.
Online Resource 4. Fifty percent majority rule consensus tree resulting from Bayesian inference of the plastid ndhF-rpl32 dataset.
Online Resource 5. The aligned data matrix used in this study.
Rights and permissions
About this article
Cite this article
Amirahmadi, A., Kazempour-Osaloo, S., Kaveh, A. et al. The phylogeny and new classification of the genus Onobrychis (Fabaceae-Hedysareae): evidence from molecular data. Plant Syst Evol 302, 1445–1456 (2016). https://doi.org/10.1007/s00606-016-1343-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-016-1343-1