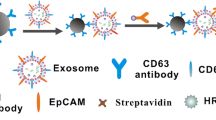
Abstract
Exosomes represent a new generation of biomarkers for the early diagnosis of hepatic carcinoma. A fluorometric assay is presented that is based on the hybridization chain reaction and magnetic nanoparticles for the highly sensitive determination of exosome (from HepG2 cells). Antibody as the recognition element was modified on the surface of magnetic nanoparticles. Antibody was used to capture the exosome. The Probe 1 was consisted of aptamer sequence and trigger sequence. Aptamer sequence will bind with the surface protein of exosome. The trigger sequence will hybridize with the probe2 (FAM-labeled) and the probe3 (FAM-labeled). So the product of the hybridization chain reaction will present a strong fluorescence signal. The fluorescence product of hybridization chain reaction will link with magnetic nanoparticles through the ‘magnetic nanoparticles-antibody-exosome-aptamer’ structure. The product can be separated from the matrix due to the present of the magnetic nanoparticles. The excitation was set at 490 nm. The fluorescence value of the emission spectra at 519 nm was set as the signal response. The linear range of this assay is from 1000 to 107 particles·mL−1. The detection limit is 100 particles·mL−1. This assay was applied to the determination of exosome from the hepatic carcinoma cells.

In the presence of exosmes, the hybridization chain reaction was triggered and strong green fluorescence will be produced. Moreover, the magnetic particles can separate the products from the matrix.





Similar content being viewed by others
References
Oliveira SD, Houseright RA, Graves AL, Golenberg N, Korte BG, Miskolci V, Huttenlocher A (2019) Metformin modulates innate immune-mediated inflammation and early progression of NAFLD-associated hepatocellular carcinoma in zebrafish. J Hepatol 70:710–721
Altekruse SF, McGlynn KA, Reichman ME (2009) Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol 27:1485–1491
Zeng J, Yin P, Tan Y, Dong L, Hu C, Huang Q, Lu X, Wang H, Xu G (2014) Metabolomics study of hepatocellular carcinoma: discovery and validation of serum potential biomarkers by using capillary electrophoresis-mass spectrometry. J Proteome Res 13:3420–3431
Rolfo C, Castiglia M, Hong D, Alessandro R, Mertens I, Baggerman G, Zwaenepoel K, Gil-Bazo I, Passiglia F, Carreca AP, Taverna S, Vento R, Santini D, Peeters M, Russo A, Pauwels P (2014) Liquid biopsies in lung cancer: the new ambrosia of researchers. Biochim Biophys Acta 1846:539–546
Lianidou E, Pantel K (2019) Liquid biopsies. Genes Chromosom Cancer 58:219–232
Buckanovich RJ, Sasaroli D, O'Brien-Jenkins A, Botbyl J, Hammond R, Katsaros D, Sandaltzopoulos R, Liotta LA, Gimotty PA, Coukos G (2007) Tumor vascular proteins as biomarkers in ovarian cancer. J Clin Oncol 25:852–861
Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, Antipova A, Lee C, McKernan K, Vega FM, Kinzler KW, Vogelstein B, Diaz JLA, Velculescu VE (2010) Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med (20):1–7
Lianidou ES, Markou A (2011) Circulating tumor cells as emerging tumor biomarkers in breast cancer. Clin Chem Lab Med 49:1579–1590
Kim JU, Shariff MI, Crossey MM, Gomez-Romero M, Holmes E, Cox IJ, Fye HK, Njie R, Taylor-Robinson SD (2016) Hepatocellular carcinoma: review of disease and tumor biomarkers. World J Hepatol 8:471–484
Thery C, Zitvogel L, Amigorena S (2002) Exosomes: composition, biogenesis and function. Nat Rev Immunol 2:569–579
Sasaki R, Kanda T, Yokosuka O, Kato N, Matsuoka S, Moriyama M (2019) Exosomes and hepatocellular carcinoma: from bench to bedside. Int J Mol Sci 20:1406
Fu M, Gu J, Jiang P, Qian H, Xu W, Zhang X (2019) Exosomes in gastric cancer: roles, mechanisms, and applications. Mol Cancer 18:41
Yao Z, Jia X, Megger DA, Chen J, Liu Y, Li J, Sitek B, Yuan Z (2019) Label-free proteomic analysis of exosomes secreted from THP-1-derived macrophages treated with IFN-alpha identifies antiviral proteins enriched in exosomes. J Proteome Res 18:855–864
Li Q, Tofaris GK, Davis JJ (2017) Concentration-normalized electroanalytical assaying of Exosomal markers. Anal Chem 89:3184–3190
Gorodkiewicz E, Lukaszewski Z (2018) Recent Progress in surface Plasmon resonance biosensors (2016 to Mid-2018). Biosensors-Basel 8:132
Xia YK, Liu MM, Wang LL, Yan A, He WH, Chen M, Lan JM, Xu JX, Guan LH, Chen JH (2017) A visible and colorimetric aptasensor based on DNA-capped single-walled carbon nanotubes for detection of exosomes. Biosens Bioelectron 92:8–15
Shin H, Jeong H, Park J, Hong S, Choi Y (2018) Correlation between cancerous exosomes and protein markers based on surface-enhanced Raman spectroscopy (SERS) and principal component analysis (PCA). ACS SENSORS 3:2637–2643
Carney RP, Hazari S, Colquhoun M, Di T, Hwang B, Mulligan MS, Bryers JD, Girda E, Leiserowitz GS, Smith ZJ, Lam KS (2017) Multispectral optical tweezers for biochemical fingerprinting of CD9-positive exosome subpopulations. Anal Chem 89:5357–5363
Busatto S, Giacomini A, Montis C, Ronca R, Bergese P (2018) Uptake profiles of human serum exosomes by murine and human tumor cells through combined use of colloidal Nanoplasmonics and flow Cytofluorimetric analysis. Anal Chem 90:7855–7861
Rupert DLM, Mapar M, Shelke GV, Norling K, Elmeskog M, Lotvall JO, Block S, Bally M, Agnarsson B, Hook F (2018) Effective refractive index and lipid content of extracellular vesicles revealed using optical waveguide scattering and fluorescence microscopy. Langmuir 34:8522–8531
Zhang K, Gan N, Hu F, Chen X, Li T, Cao J (2018) Microfluidic electrophoretic non-enzymatic kanamycin assay making use of a stirring bar functionalized with gold-labeled aptamer, of a fluorescent DNA probe, and of signal amplification via hybridization chain reaction. Microchim Acta 185:181
Keefe AD, Pai S, Ellington A (2010) Aptamers as therapeutics. Nat Rev Drug Discov 9:537–550
Hong M, Wang M, Wang J, Xu X, Lin Z (2017) Ultrasensitive and selective electrochemical biosensor for detection of mercury (II) ions by nicking endonuclease-assisted target recycling and hybridization chain reaction signal amplification. Biosens Bioelectron 94:19–23
Yin J, Gan P, Zhou F, Wang J (2014) Sensitive detection of transcription factors using near-infrared fluorescent solid-phase rolling circle amplification. Anal Chem 86:2572–2579
Shen R, Zou L, Wu S, Li T, Wang J, Liu J, Ling L (2019) A novel label-free fluorescent detection of histidine based upon Cu(2+)-specific DNAzyme and hybridization chain reaction. Spectrochim Acta A Mol Biomol Spectrosc 213:42–47
Wu C, Cansiz S, Zhang L, Teng IT, Qiu L, Li J, Liu Y, Zhou C, Hu R, Zhang T, Cui C, Cui L, Tan W (2015) A nonenzymatic hairpin DNA Cascade reaction provides high signal gain of mRNA imaging inside live cells. J Am Chem Soc 137:4900–4903
Lu AH, Salabas EL, Schuth F (2007) Magnetic nanoparticles: synthesis, protection, functionalization, and application. Angew Chem 46:1222–1244
Hao R, Xing R, Xu Z, Hou Y, Gao S, Sun S (2010) Synthesis, functionalization, and biomedical applications of multifunctional magnetic nanoparticles. Adv Mater 22:2729–2742
Yang Y, Li C, Shi H, Chen T, Wang Z, Li G (2019) A pH-responsive bioassay for paper-based diagnosis of exosomes via mussel-inspired surface chemistry. Talanta 192:325–330
Tian YF, Ning CF, He F, Yin BC, Ye BC (2018) Highly sensitive detection of exosomes by SERS using gold nanostar@ Raman reporter@ nanoshell structures modified with a bivalent cholesterollabeled DNA anchor. Analyst 143(20):4915–4922
Zhu L, Wang K, Cui J, Liu H, Bu X, Ma H, Wang W, Gong H, Lausted C, Hood L et al (2014) Label-free quantitative detection of tumor-derived exosomes through surface Plasmon resonance imaging. Anal Chem 86(17):8857–8864
Xu H, Liao C, Zuo P, Liu Z, Ye B-C (2018) Magnetic-based microfluidic device for on-Chip isolation and detection of tumor-derived exosomes. Anal Chem 90(22):13451–13458
Zhang Q, Wang F, Zhang H, Zhang Y, Liu M, Liu Y (2018) Universal Ti3C2 MXenes based self-standard Ratiometric fluorescence resonance energy transfer platform for highly sensitive detection of exosomes. Anal Chem 90(21):12737–12744
Boriachek K, Masud MK, Palma C, Phan HP, Yamauchi Y, Hossain MSA, Nguyen NT, Salomon C, Shiddiky MJA (2019) Avoiding pre-isolation step in exosome analysis: direct isolation and sensitive detection of exosomes using gold-loaded Nanoporous ferric oxide Nanozymes. Anal Chem 91(6):3827–3834
Acknowledgements
This work was supported by Hubei Natural Science Fund (2015CFB668) and Independent Innovation Fund of Huazhong University of Science and Technology (0118530318).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 387 kb)
Rights and permissions
About this article
Cite this article
Shi, L., Ba, L., Xiong, Y. et al. A hybridization chain reaction based assay for fluorometric determination of exosomes using magnetic nanoparticles and both aptamers and antibody as recognition elements. Microchim Acta 186, 796 (2019). https://doi.org/10.1007/s00604-019-3823-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-3823-9